Induction of autophagy by second-fermentation yeasts during elaboration of sparkling wines
- PMID: 16751523
- PMCID: PMC1489611
- DOI: 10.1128/AEM.02920-05
Induction of autophagy by second-fermentation yeasts during elaboration of sparkling wines
Abstract
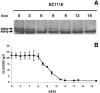
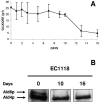
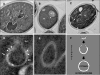
Autophagy is a transport system mediated by vesicles, ubiquitous in eukaryotic cells, by which bulk cytoplasm is targeted to a lysosome or vacuole for degradation. In the yeast Saccharomyces cerevisiae, autophagy is triggered by nutritional stress conditions (e.g., carbon- or nitrogen-depleted medium). In this study we showed that there is induction of autophagy in second-fermentation yeasts during sparkling wine making. Two methods were employed to detect autophagy: a biochemical approach based on depletion of the protein acetaldehyde dehydrogenase Ald6p and a morphological strategy consisting of visualization of autophagic bodies and autophagosomes, which are intermediate vesicles in the autophagic process, by transmission electron microscopy. This study provides the first demonstration of autophagy in second-fermentation yeasts under enological conditions. The correlation between autophagy and yeast autolysis during sparkling wine production is discussed, and genetic engineering of autophagy-related genes in order to accelerate the aging steps in wine making is proposed.
Figures



Similar articles
-
Autophagy in wine making.Methods Enzymol. 2008;451:163-75. doi: 10.1016/S0076-6879(08)03212-6. Methods Enzymol. 2008. PMID: 19185720
-
Evidence for yeast autophagy during simulation of sparkling wine aging: a reappraisal of the mechanism of yeast autolysis in wine.Biotechnol Prog. 2005 Mar-Apr;21(2):614-6. doi: 10.1021/bp049708y. Biotechnol Prog. 2005. PMID: 15801807
-
Sensory and analytical study of rose sparkling wines manufactured by second fermentation in the bottle.J Agric Food Chem. 2004 Oct 20;52(21):6640-5. doi: 10.1021/jf040151b. J Agric Food Chem. 2004. PMID: 15479034
-
Enological functions of parietal yeast mannoproteins.Antonie Van Leeuwenhoek. 2006 Apr-May;89(3-4):417-22. doi: 10.1007/s10482-005-9050-x. Epub 2006 Apr 19. Antonie Van Leeuwenhoek. 2006. PMID: 16622788 Review.
-
[Physiological and biochemical characteristics of immobilized champagne yeasts and their participation in sparkling processes].Prikl Biokhim Mikrobiol. 2003 Sep-Oct;39(5):501-8. Prikl Biokhim Mikrobiol. 2003. PMID: 14593861 Review. Russian.
Cited by
-
The Transcriptomic Mechanism of a Novel Autolysis Induced by a Recombinant Antibacterial Peptide from Chicken Expressed in Pichia pastoris.Molecules. 2022 Mar 21;27(6):2029. doi: 10.3390/molecules27062029. Molecules. 2022. PMID: 35335392 Free PMC article.
-
The multiple roles of lipid metabolism in yeast physiology during beer fermentation.Genet Mol Biol. 2022 Sep 16;45(3):e20210325. doi: 10.1590/1678-4685-GMB-2021-0325. eCollection 2022. Genet Mol Biol. 2022. PMID: 36149459 Free PMC article.
-
Autolysis of Pichia pastoris induced by cold.AMB Express. 2017 Dec;7(1):95. doi: 10.1186/s13568-017-0397-y. Epub 2017 May 12. AMB Express. 2017. PMID: 28500590 Free PMC article.
-
Quantitative Data-Independent Acquisition Glycoproteomics of Sparkling Wine.Mol Cell Proteomics. 2021;20:100020. doi: 10.1074/mcp.RA120.002181. Epub 2020 Dec 21. Mol Cell Proteomics. 2021. PMID: 32938748 Free PMC article.
-
Biological Processes Highlighted in Saccharomyces cerevisiae during the Sparkling Wines Elaboration.Microorganisms. 2020 Aug 11;8(8):1216. doi: 10.3390/microorganisms8081216. Microorganisms. 2020. PMID: 32796563 Free PMC article.
References
-
- Andrés-Lacueva, C., R. M. Lamuela-Raventós, S. Buxaderas, and M. C. De la Torre-Boronat. 1997. Influence of variety and aging on foaming properties of cava (sparkling wine). J. Agric. Food Chem. 45:2520-2525.
-
- Bernfeld, P. 1955. Amylases α and β. Methods Enzymol. 1:149-158.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

