Molecular architecture of E. coli purine nucleoside phosphorylase studied by analytical ultracentrifugation and CD spectroscopy
- PMID: 16751611
- PMCID: PMC2242567
- DOI: 10.1110/ps.062183206
Molecular architecture of E. coli purine nucleoside phosphorylase studied by analytical ultracentrifugation and CD spectroscopy
Abstract
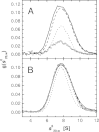
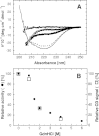
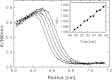
Purine nucleoside phosphorylase (PNP) is a key enzyme of the nucleoside salvage pathway and is characterized by complex kinetics. It was suggested that this is due to coexistence of various oligomeric forms that differ in specific activity. In this work, the molecular architecture of Escherichia coli PNP in solution was studied by analytical ultracentrifugation and CD spectroscopy. Sedimentation equilibrium analysis revealed a homohexameric molecule with molecular mass 150+/-10 kDa, regardless of the conditions investigated-protein concentration, 0.18-1.7 mg/mL; presence of up to 10 mM phosphate and up to 100 mM KCl; temperature, 4-20 degrees C. The parameters obtained from the self-associating model also describe the hexameric form. Sedimentation velocity experiments conducted for broad protein concentration range (1 microg/mL-1.3 mg/mL) with boundary (classical) and band (active enzyme) approaches gave s(0)20,w=7.7+/-0.3 and 8.3+/-0.4 S, respectively. The molecular mass of the sedimenting particle (146+/-30 kDa), calculated using the Svedberg equation, corresponds to the mass of the hexamer. Relative values of the CD signal at 220 nm and the catalytic activity of PNP as a function of GdnHCl concentration were found to be correlated. The transition from the native state to the random coil is a single-step process. The sedimentation coefficient determined at 1 M GdnHCl (at which the enzyme is still fully active) is 7.7 S, showing that also under these conditions the hexamer is the only catalytically active form. Hence, in solution similar to the crystal, E. coli PNP is a hexameric molecule and previous suggestions for coexistence of two oligomeric forms are incorrect.
Figures



References
-
- Behlke J., Koellner G., Bzowska A. 2005. Oligomeric structure of mammalian purine nucleoside phosphorylase in solution determined by analytical ultracentrifugation. Z. Naturforsch. 60C 927–931. - PubMed
-
- Bzowska A. 2002. Calf spleen purine nucleoside phosphorylase: Complex kinetic mechanism, hydrolysis of 7-methylguanosine, and oligomeric state in solution. Biochim. Biophys. Acta 1596 293–317. - PubMed
-
- Bzowska A., Kazimierczuk Z., Seela F. 1998. 7-Deazapurine 2′-deoxyribofuranosides are noncleavable competitive inhibitors of Escherichia coli purine nucleoside phosphorylase (PNP). Acta Biochim. Pol. 45 755–768. - PubMed
-
- Bzowska A., Kulikowska E., Shugar D. 2000. Purine nucleoside phosphorylases: Properties, functions, and clinical aspects. Pharmacol. Ther. 88 349–425. - PubMed
-
- Cohen R. and Claverie J.-M. 1975. Sedimentation of generalized systems of interacting particles. II. Active enzyme centrifugation—theory and extensions of its validity range. Biopolymers 14 1701–1716. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

