Deletion polymorphism of Disc1 is common to all 129 mouse substrains: implications for gene-targeting studies of brain function
- PMID: 16751659
- PMCID: PMC1569715
- DOI: 10.1534/genetics.106.060749
Deletion polymorphism of Disc1 is common to all 129 mouse substrains: implications for gene-targeting studies of brain function
Abstract
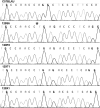
We report that the Disc1 gene in all extant 129 mouse inbred substrains has a deletion, previously considered specific to the 129S6/SvEv substrain, which is predicted to abolish production of the full-length protein. This finding has implications for the study of knockout mice generated from 129-derived embryonic stem cells.
Figures
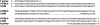

References
-
- Beck, J. A., S. Lloyd, M. Hafezparast, M. Lennon-Pierce, J. T. Eppig et al., 2000. Genealogies of mouse inbred strains. Nat. Genet. 24: 23–25. - PubMed
-
- Bjorklund, M., I. Siverina, T. Heikkinen, H. Tanila, J. Sallinen et al., 2001. Spatial working memory improvement by an alpha2-adrenoceptor agonist dexmedetomidine is not mediated through alpha2C-adrenoceptor. Prog. Neuropsychopharmacol. Biol. Psychiatry 25: 1539–1554. - PubMed
-
- Cannon, T. D., W. Hennah, T. G. M. van Erp, P. M. Thompson, J. Lonnqvist et al., 2005. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch. Gen. Psychiatry 62: 1205–1213. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

