Long-term evolution and functional diversification in the members of the nucleophosmin/nucleoplasmin family of nuclear chaperones
- PMID: 16751661
- PMCID: PMC1569712
- DOI: 10.1534/genetics.106.058990
Long-term evolution and functional diversification in the members of the nucleophosmin/nucleoplasmin family of nuclear chaperones
Abstract
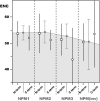
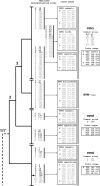
The proper assembly of basic proteins with nucleic acids is a reaction that must be facilitated to prevent protein aggregation and formation of nonspecific nucleoprotein complexes. The proteins that mediate this orderly protein assembly are generally termed molecular (or nuclear) chaperones. The nucleophosmin/nucleoplasmin (NPM) family of molecular chaperones encompasses members ubiquitously expressed in many somatic tissues (NPM1 and -3) or specific to oocytes and eggs (NPM2). The study of this family of molecular chaperones has experienced a renewed interest in the past few years. However, there is a lack of information regarding the molecular evolution of these proteins. This work represents the first attempt to characterize the long-term evolution followed by the members of this family. Our analysis shows that there is extensive silent divergence at the nucleotide level suggesting that this family has been subject to strong purifying selection at the protein level. In contrast to NPM1 and NPM-like proteins in invertebrates, NPM2 and NPM3 have a polyphyletic origin. Furthermore, the presence of selection for high frequencies of acidic residues as well as the existence of higher levels of codon bias was detected at the C-terminal ends, which can be ascribed to the critical role played by these residues in constituting the acidic tracts and to the preferred codon usage for phosphorylatable amino acids at these regions.
Figures





References
-
- Arnan, C., N. Saperas, C. Prieto, M. Chiva and J. Ausió, 2003. Interaction of nucleoplasmin with core histones. J. Biol. Chem. 278: 31319–31324. - PubMed
-
- Bañuelos, S., A. Hierro, J. M. Arizmendi, G. Montoya, A. Prado et al., 2003. Activation mechanism of the nuclear chaperone nucleoplasmin: role of the core domain. J. Mol. Biol. 334: 585–593. - PubMed
-
- Blom, N., S. Gammeltoft and S. Brunak, 1999. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294: 1351–1362. - PubMed
-
- Borer, R. A., C. F. Lehner, H. M. Eppenberger and E. A. Nigg, 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56: 379–390. - PubMed
-
- Burns, K. H., M. M. Viveiros, Y. Ren, P. Wang, F. J. DeMayo et al., 2003. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science 300: 633–636. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

