The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae
- PMID: 16751663
- PMCID: PMC1698614
- DOI: 10.1534/genetics.106.057836
The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae
Abstract
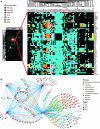
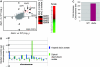
The maintenance of DNA replication fork stability under conditions of DNA damage and at natural replication pause sites is essential for genome stability. Here, we describe a novel role for the F-box protein Dia2 in promoting genome stability in the budding yeast Saccharomyces cerevisiae. Like most other F-box proteins, Dia2 forms a Skp1-Cdc53/Cullin-F-box (SCF) E3 ubiquitin-ligase complex. Systematic analysis of genetic interactions between dia2Delta and approximately 4400 viable gene deletion mutants revealed synthetic lethal/synthetic sick interactions with a broad spectrum of DNA replication, recombination, checkpoint, and chromatin-remodeling pathways. dia2Delta strains exhibit constitutive activation of the checkpoint kinase Rad53 and elevated counts of endogenous DNA repair foci and are unable to overcome MMS-induced replicative stress. Notably, dia2Delta strains display a high rate of gross chromosomal rearrangements (GCRs) that involve the rDNA locus and an increase in extrachromosomal rDNA circle (ERC) formation, consistent with an observed enrichment of Dia2 in the nucleolus. These results suggest that Dia2 is essential for stable passage of replication forks through regions of damaged DNA and natural fragile regions, particularly the replication fork barrier (RFB) of rDNA repeat loci. We propose that the SCFDia2 ubiquitin ligase serves to modify or degrade protein substrates that would otherwise impede the replication fork in problematic regions of the genome.
Figures






References
-
- Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler et al., 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965. - PubMed
-
- Araki, Y., Y. Kawasaki, H. Sasanuma, B. K. Tye and A. Sugino, 2003. Budding yeast mcm10/dna43 mutant requires a novel repair pathway for viability. Genes Cells 8: 465–480. - PubMed
-
- Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl et al., 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

