Why do hubs tend to be essential in protein networks?
- PMID: 16751849
- PMCID: PMC1473040
- DOI: 10.1371/journal.pgen.0020088
Why do hubs tend to be essential in protein networks?
Abstract
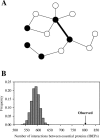
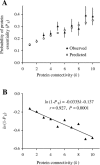
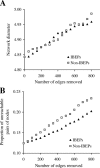
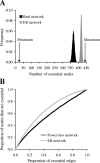
The protein-protein interaction (PPI) network has a small number of highly connected protein nodes (known as hubs) and many poorly connected nodes. Genome-wide studies show that deletion of a hub protein is more likely to be lethal than deletion of a non-hub protein, a phenomenon known as the centrality-lethality rule. This rule is widely believed to reflect the special importance of hubs in organizing the network, which in turn suggests the biological significance of network architectures, a key notion of systems biology. Despite the popularity of this explanation, the underlying cause of the centrality-lethality rule has never been critically examined. We here propose the concept of essential PPIs, which are PPIs that are indispensable for the survival or reproduction of an organism. Our network analysis suggests that the centrality-lethality rule is unrelated to the network architecture, but is explained by the simple fact that hubs have large numbers of PPIs, therefore high probabilities of engaging in essential PPIs. We estimate that approximately 3% of PPIs are essential in the yeast, accounting for approximately 43% of essential genes. As expected, essential PPIs are evolutionarily more conserved than nonessential PPIs. Considering the role of essential PPIs in determining gene essentiality, we find the yeast PPI network functionally more robust than random networks, yet far less robust than the potential optimum. These and other findings provide new perspectives on the biological relevance of network structure and robustness.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
References
-
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. - PubMed
-
- Newman MEJ. The structure and function of complex networks. SIAM Review. 2003;45:167–256.
-
- Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. - PubMed
-
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. - PubMed
-
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

