Expression and functional characterization of the putative protein 8b of the severe acute respiratory syndrome-associated coronavirus
- PMID: 16753150
- PMCID: PMC7094570
- DOI: 10.1016/j.febslet.2006.05.051
Expression and functional characterization of the putative protein 8b of the severe acute respiratory syndrome-associated coronavirus
Abstract
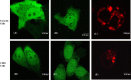
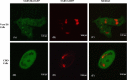
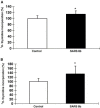
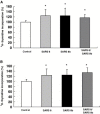
SARS 8b is one of the putative accessory proteins of the severe acute respiratory syndrome-associated coronavirus (SARS-CoV) with unknown functions. In this study, the cellular localization and activity of this estimated 9.6 kDa protein were examined. Confocal microscopy results indicated that SARS 8b is localized in both nucleus and cytoplasm of mammalian cells. Functional study revealed that overexpression of SARS 8b induced DNA synthesis. Coexpression of SARS 8b and SARS 6, a previously characterized SARS-CoV accessory protein, did not elicit synergistic effects on DNA synthesis.
Figures


Similar articles
-
The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells.Virology. 2006 Oct 10;354(1):132-42. doi: 10.1016/j.virol.2006.06.026. Epub 2006 Jul 31. Virology. 2006. PMID: 16876844 Free PMC article.
-
The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization.FEBS Lett. 2005 Dec 19;579(30):6763-8. doi: 10.1016/j.febslet.2005.11.007. Epub 2005 Nov 21. FEBS Lett. 2005. PMID: 16310783 Free PMC article.
-
Expression, post-translational modification and biochemical characterization of proteins encoded by subgenomic mRNA8 of the severe acute respiratory syndrome coronavirus.FEBS J. 2007 Aug;274(16):4211-22. doi: 10.1111/j.1742-4658.2007.05947.x. Epub 2007 Jul 20. FEBS J. 2007. PMID: 17645546 Free PMC article.
-
SARS coronavirus accessory proteins.Virus Res. 2008 Apr;133(1):113-21. doi: 10.1016/j.virusres.2007.10.009. Epub 2007 Nov 28. Virus Res. 2008. PMID: 18045721 Free PMC article. Review.
-
Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus.Antiviral Res. 2006 Nov;72(2):78-88. doi: 10.1016/j.antiviral.2006.05.010. Epub 2006 Jun 6. Antiviral Res. 2006. PMID: 16820226 Free PMC article. Review.
Cited by
-
SARS-CoV-2 mutations in Brazil: from genomics to putative clinical conditions.Sci Rep. 2021 Jun 7;11(1):11998. doi: 10.1038/s41598-021-91585-6. Sci Rep. 2021. PMID: 34099808 Free PMC article.
-
Product of natural evolution (SARS, MERS, and SARS-CoV-2); deadly diseases, from SARS to SARS-CoV-2.Hum Vaccin Immunother. 2021 Jan 2;17(1):62-83. doi: 10.1080/21645515.2020.1797369. Epub 2020 Aug 12. Hum Vaccin Immunother. 2021. PMID: 32783700 Free PMC article.
-
Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination.J Virol. 2015 Oct;89(20):10532-47. doi: 10.1128/JVI.01048-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269185 Free PMC article.
-
Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission.Sci Rep. 2018 Oct 11;8(1):15177. doi: 10.1038/s41598-018-33487-8. Sci Rep. 2018. PMID: 30310104 Free PMC article.
-
SARS-CoV-2: Insights into its structural intricacies and functional aspects for drug and vaccine development.Int J Biol Macromol. 2021 May 15;179:45-60. doi: 10.1016/j.ijbiomac.2021.02.212. Epub 2021 Mar 1. Int J Biol Macromol. 2021. PMID: 33662418 Free PMC article. Review.
References
-
- World Health Organization (WHO). http://www.who.int/csr/sars/en/.
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science, 300, (2003), 1394– 1399. - PubMed
-
- Marra M.A., Jones S.J.M., Astell C.R., The genome sequence of the SARS-associated coronavirus. Science, 300, (2003), 1399– 1404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

