Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress
- PMID: 16754955
- PMCID: PMC2063885
- DOI: 10.1083/jcb.200602108
Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress
Abstract
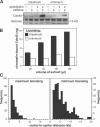
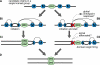
In late mitosis and early G1, replication origins are licensed for subsequent use by loading complexes of the minichromosome maintenance proteins 2-7 (Mcm2-7). The number of Mcm2-7 complexes loaded onto DNA greatly exceeds the number of replication origins used during S phase, but the function of the excess Mcm2-7 is unknown. Using Xenopus laevis egg extracts, we show that these excess Mcm2-7 complexes license additional dormant origins that do not fire during unperturbed S phases because of suppression by a caffeine-sensitive checkpoint pathway. Use of these additional origins can allow complete genome replication in the presence of replication inhibitors. These results suggest that metazoan replication origins are actually comprised of several candidate origins, most of which normally remain dormant unless cells experience replicative stress. Consistent with this model, using Caenorhabditis elegans, we show that partial RNAi-based knockdown of MCMs that has no observable effect under normal conditions causes lethality upon treatment with low, otherwise nontoxic, levels of the replication inhibitor hydroxyurea.
Figures





References
-
- Alexandrow, M.G., M. Ritzi, A. Pemov, and J.L. Hamlin. 2002. A potential role for mini-chromosome maintenance (MCM) proteins in initiation at the dihydrofolate reductase replication origin. J. Biol. Chem. 277:2702–2708. - PubMed
-
- Anglana, M., F. Apiou, A. Bensimon, and M. Debatisse. 2003. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 114:385–394. - PubMed
-
- Aparicio, O.M., D.M. Weinstein, and S.P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 91:59–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

