Purification, crystallization and preliminary X-ray analysis of glutathione peroxidase Gpx3 from Saccharomyces cerevisiae
- PMID: 16754991
- PMCID: PMC2243105
- DOI: 10.1107/S1744309106017829
Purification, crystallization and preliminary X-ray analysis of glutathione peroxidase Gpx3 from Saccharomyces cerevisiae
Abstract
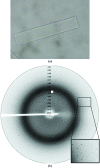
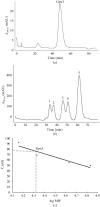
The glutathione peroxidase Gpx3 from the yeast Saccharomyces cerevisiae has been overexpressed, purified and crystallized. Both gel-filtration and dynamic light-scattering (DLS) results indicate that Gpx3 is a monomer in solution at a concentration of about 2 mg ml(-1), whereas glutathione peroxidases are normally tetrameric or dimeric. X-ray diffraction data from a single crystal of Gpx3 have been collected to 2.6 A resolution. The crystals are triclinic and belong to space group P1, with unit-cell parameters a = 38.187, b = 43.372, c = 56.870 A, alpha = 71.405, beta = 73.376, gamma = 89.633 degrees. There are two Gpx3 monomers in a crystallographic asymmetric unit. Preliminary analyses show that the yeast Gpx3 is quite different from those of mammals.
Figures


References
-
- Avery, A. M. & Avery, S. V. (2001). J. Biol. Chem.276, 33730–33735. - PubMed
-
- Basu, U., Southron, J. L., Stephens, J. L. & Taylor, G. J. (2004). Mol. Genet. Genomics, 271, 627–637. - PubMed
-
- Brigelius-Flohe, R., Aumann, K. D., Blocker, H., Gross, G., Kiess, M., Kloppel, K. D., Maiorino, M., Roveri, A., Schuckelt, R., Ursini, F., Wingender, E. & Flohe, L. (1994). J. Biol. Chem.269, 7342–7348. - PubMed
-
- Claiborne, A., Yeh, J. I., Mallett, T. C., Luba, J., Crane, E. J., Charrier, V. & Parsonage, D. (1999). Biochemistry, 38, 15407–15416. - PubMed
-
- Delaunay, A., Pflieger, D., Barrault, M. B., Vinh, J. & Toledano, M. B. (2002). Cell, 111, 471–481. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

