DNA detection using recombination proteins
- PMID: 16756388
- PMCID: PMC1475771
- DOI: 10.1371/journal.pbio.0040204
DNA detection using recombination proteins
Abstract
DNA amplification is essential to most nucleic acid testing strategies, but established techniques require sophisticated equipment or complex experimental procedures, and their uptake outside specialised laboratories has been limited. Our novel approach, recombinase polymerase amplification (RPA), couples isothermal recombinase-driven primer targeting of template material with strand-displacement DNA synthesis. It achieves exponential amplification with no need for pretreatment of sample DNA. Reactions are sensitive, specific, and rapid and operate at constant low temperature. We have also developed a probe-based detection system. Key aspects of the combined RPA amplification/detection process are illustrated by a test for the pathogen methicillin-resistant Staphylococcus aureus. The technology proves to be sensitive to fewer than ten copies of genomic DNA. Furthermore, products can be detected in a simple sandwich assay, thereby establishing an instrument-free DNA testing system. This unique combination of properties is a significant advance in the development of portable and widely accessible nucleic acid-based tests.
Figures
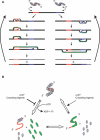
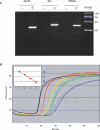


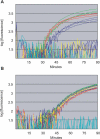

Comment in
-
DNA amplification and detection made simple (relatively).PLoS Biol. 2006 Jul;4(7):e222. doi: 10.1371/journal.pbio.0040222. Epub 2006 Jun 13. PLoS Biol. 2006. PMID: 20076600 Free PMC article. No abstract available.
References
-
- Yonesaki T, Ryo Y, Minagawa T, Takahashi H. Purification and some of the functions of the products of bacteriophage T4 recombination genes, uvsX and uvsY. Eur J Biochem. 1985;148:127–134. - PubMed
-
- Formosa T, Alberts BM. Purification and characterization of the T4 bacteriophage uvsX protein. J Biol Chem. 1986;261:6107–6118. - PubMed
-
- Harris LD, Griffith JD. Formation of D loops by the UvsX protein of T4 bacteriophage: A comparison of the reaction catalyzed in the presence or absence of gene 32 protein. Biochemistry. 1988;27:6954–6959. - PubMed
-
- Okazaki T, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. Xv.purification and properties of a polymerase from Bacillus subtilis . J Biol Chem. 1964;239:259–268. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

