The bacterial twin-arginine translocation pathway
- PMID: 16756481
- PMCID: PMC2654714
- DOI: 10.1146/annurev.micro.60.080805.142212
The bacterial twin-arginine translocation pathway
Abstract
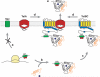
The twin-arginine translocation (Tat) pathway is responsible for the export of folded proteins across the cytoplasmic membrane of bacteria. Substrates for the Tat pathway include redox enzymes requiring cofactor insertion in the cytoplasm, multimeric proteins that have to assemble into a complex prior to export, certain membrane proteins, and proteins whose folding is incompatible with Sec export. These proteins are involved in a diverse range of cellular activities including anaerobic metabolism, cell envelope biogenesis, metal acquisition and detoxification, and virulence. The Escherichia coli translocase consists of the TatA, TatB, and TatC proteins, but little is known about the precise sequence of events that leads to protein translocation, the energetic requirements, or the mechanism that prevents the export of misfolded proteins. Owing to the unique characteristics of the pathway, it holds promise for biotechnological applications.
Figures



References
-
- Alami M, Luke I, Deitermann S, Eisner G, Koch HG, et al. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell. 2003;12:937–46. - PubMed
-
- Alami M, Trescher D, Wu LF, Muller M. Separate analysis of twin-arginine translocation (Tat)-specific membrane binding and translocation in Escherichia coli. J. Biol. Chem. 2002;277:20499–503. - PubMed
-
- Alder NN, Theg SM. Energetics of protein transport across biological membranes. A study of the thylakoid DeltapH-dependent/cpTat pathway. Cell. 2003;112:231–42. - PubMed
-
- Alder NN, Theg SM. Energy use by biological protein transport pathways. Trends Biochem. Sci. 2003;28:442–51. - PubMed
-
- Allen SC, Barrett CM, Ray N, Robinson C. Essential cytoplasmic domains in the Escherichia coli TatC protein. J. Biol. Chem. 2002;277:10362–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

