Characteristics of MuA transposase-catalyzed processing of model transposon end DNA hairpin substrates
- PMID: 16757579
- PMCID: PMC1475752
- DOI: 10.1093/nar/gkl405
Characteristics of MuA transposase-catalyzed processing of model transposon end DNA hairpin substrates
Abstract
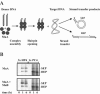
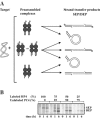
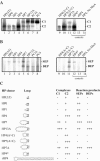
Bacteriophage Mu uses non-replicative transposition for integration into the host's chromosome and replicative transposition for phage propagation. Biochemical and structural comparisons together with evolutionary considerations suggest that the Mu transposition machinery might share functional similarities with machineries of the systems that are known to employ a hairpin intermediate during the catalytic steps of transposition. Model transposon end DNA hairpin substrates were used in a minimal-component in vitro system to study their proficiency to promote Mu transpososome assembly and subsequent MuA-catalyzed chemical reactions leading to the strand transfer product. MuA indeed was able to assemble hairpin substrates into a catalytically competent transpososome, open the hairpin ends and accurately join the opened ends to the target DNA. The hairpin opening and transposon end cleavage reactions had identical metal ion preferences, indicating similar conformations within the catalytic center for these reactions. Hairpin length influenced transpososome assembly as well as catalysis: longer loops were more efficient in these respects. In general, MuA's proficiency to utilize different types of hairpin substrates indicates a certain degree of flexibility within the transposition machinery core. Overall, the results suggest that non-replicative and replicative transposition systems may structurally and evolutionarily be more closely linked than anticipated previously.
Figures

Similar articles
-
Mechanisms of metal ion action in Tn10 transposition.J Mol Biol. 2002 May 24;319(1):53-65. doi: 10.1016/S0022-2836(02)00297-8. J Mol Biol. 2002. PMID: 12051936
-
Functional comparison of the transposition core machineries of phage Mu and Haemophilus influenzae Mu-like prophage Hin-Mu reveals interchangeable components.Virology. 2005 Jan 5;331(1):6-19. doi: 10.1016/j.virol.2004.09.041. Virology. 2005. PMID: 15582649
-
3D reconstruction of the Mu transposase and the Type 1 transpososome: a structural framework for Mu DNA transposition.Genes Dev. 2005 Apr 1;19(7):840-52. doi: 10.1101/gad.1291405. Epub 2005 Mar 17. Genes Dev. 2005. PMID: 15774720 Free PMC article.
-
[Transposition as a way of existence: phage Mu].Genetika. 2003 May;39(5):637-56. Genetika. 2003. PMID: 12838611 Review. Russian.
-
Structure/function insights into Tn5 transposition.Curr Opin Struct Biol. 2004 Feb;14(1):50-7. doi: 10.1016/j.sbi.2004.01.008. Curr Opin Struct Biol. 2004. PMID: 15102449 Review.
Cited by
-
Transposable Phage Mu.Microbiol Spectr. 2014 Oct;2(5):10.1128/microbiolspec.MDNA3-0007-2014. doi: 10.1128/microbiolspec.MDNA3-0007-2014. Microbiol Spectr. 2014. PMID: 26104374 Free PMC article. Review.
-
Flexibility in MuA transposase family protein structures: functional mapping with scanning mutagenesis and sequence alignment of protein homologues.PLoS One. 2012;7(5):e37922. doi: 10.1371/journal.pone.0037922. Epub 2012 May 29. PLoS One. 2012. PMID: 22666413 Free PMC article.
-
Alternative mechanisms for tn5 transposition.PLoS Genet. 2009 Aug;5(8):e1000619. doi: 10.1371/journal.pgen.1000619. Epub 2009 Aug 28. PLoS Genet. 2009. PMID: 19714209 Free PMC article.
-
Nanopore sequencing of native adeno-associated virus (AAV) single-stranded DNA using a transposase-based rapid protocol.NAR Genom Bioinform. 2020 Sep 28;2(4):lqaa074. doi: 10.1093/nargab/lqaa074. eCollection 2020 Dec. NAR Genom Bioinform. 2020. PMID: 33575623 Free PMC article.
-
Thorough molecular configuration analysis of noncanonical AAV genomes in AAV vector preparations.Mol Ther Methods Clin Dev. 2024 Feb 19;32(1):101215. doi: 10.1016/j.omtm.2024.101215. eCollection 2024 Mar 14. Mol Ther Methods Clin Dev. 2024. PMID: 38463141 Free PMC article.
References
-
- Craig N.L., Craigie R., Gellert M., Lambowitz A.M. (eds) Mobile DNA II. Washington DC: American Society for Microbiology; 2002.
-
- Curcio J.M., Derbyshire K.M. The outs and ins of transposition: from Mu to kangaroo. Mol. Cell. Biol. 2003;4:1–13. - PubMed
-
- Craig N. Unity in transposition reactions. Science. 1995;270:253–254. - PubMed
-
- Gueguen E., Rousseau P., Duval-Valentin G., Chandler M. The transpososome: control of transposition at the level of catalysis. Trends Microbiol. 2005;13:543–549. - PubMed
-
- Rice P., Mizuuchi K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition. Cell. 1995;82:209–220. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

