Identification of a Haarlem genotype-specific single nucleotide polymorphism in the mgtC virulence gene of Mycobacterium tuberculosis
- PMID: 16757603
- PMCID: PMC1489410
- DOI: 10.1128/JCM.00278-06
Identification of a Haarlem genotype-specific single nucleotide polymorphism in the mgtC virulence gene of Mycobacterium tuberculosis
Erratum in
- J Clin Microbiol. 2008 Sep;46(9):3183
Abstract
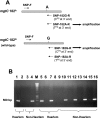
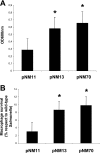
MgtC is a virulence factor common to several intracellular pathogens, including Mycobacterium tuberculosis, that might have been acquired through horizontal gene transfer. In the present study, we investigated the polymorphism of mgtC in clinical isolates representative of the main epidemic groups of M. tuberculosis. MgtC appears to have a low polymorphism rate in M. tuberculosis that consists exclusively of nonsynonymous mutations. We identified a single nucleotide polymorphism (SNP) at mgtC codon 182 (mgtC182) specifically associated with the Haarlem genotype. A simple PCR assay, called the "on/off switch assay," using phosphorothioate-modified primers and Pfu polymerase allowed us to distinguish Haarlem from non-Haarlem strains based on the mgtC182 SNP. The amino acid change (H182R) associated with the mgtC182 SNP in Haarlem strains does not appear to procure a selective advantage. Our results offer a simple and rapid tool to distinguish between Haarlem and non-Haarlem strains. In addition, the on/off switch assay, which allows the detection of SNPs on chromosomal DNA and M. tuberculosis cultures, provides a novel approach for the screening of known SNPs in M. tuberculosis.
Figures
References
-
- Blanc-Potard, A. B., and B. Lafay. 2003. MgtC as a horizontally acquired virulence factor of intracellular bacterial pathogens: evidence from molecular phylogeny and comparative genomics. J. Mol. Evol. 57:479-486. - PubMed
-
- Buchmeier, N., A. Blanc-Potard, S. Ehrt, D. Piddington, L. Riley, and E. A. Groisman. 2000. A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol. Microbiol. 35:1375-1382. - PubMed
-
- Cave, M. D., K. D. Eisenach, P. F. McDermott, J. H. Bates, and J. T. Crawford. 1991. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol. Cell Probes 5:73-80. - PubMed
-
- Dolzani, L., M. Rosato, B. Sartori, E. Banfi, C. Lagatolla, M. Predominato, C. Fabris, E. Tonin, F. Gombac, and C. Monti-Bragadin. 2004. Mycobacterium tuberculosis isolates belonging to katG gyrA group 2 are associated with clustered cases of tuberculosis in Italian patients. J. Med. Microbiol. 53:155-159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

