Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from Eastern and Western Canada
- PMID: 16757612
- PMCID: PMC1489423
- DOI: 10.1128/JCM.02563-05
Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from Eastern and Western Canada
Abstract
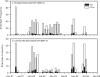
The prevalence and characteristics of PCR ribotype 027 strains of Clostridium difficile have come into question following recent outbreaks in Eastern Canada and elsewhere. In order to determine the distribution of this strain in other regions in Canada, we screened a bank of 1,419 isolates recovered from three different Canadian health regions between 2000 and 2004. Among isolates from a Montreal area hospital, PCR ribotype 027 strains represented 115/153 strains (75.2%) from 2003 to 2004, but ribotype 027 strains were absent in 2000 and 2001. In Calgary, by contrast, ribotype 027 rates have remained relatively stable over 4 years of surveillance, representing 51/685 (7.4%) hospital isolates and 62/373 (16.6%) strains from the community (P < 0.001). PCR ribotype 027 accounted for 8/135 (5.9%) hospital isolates in the Fraser Health Region in 2004. repetitive extragenic palindromic PCR was used to subtype a random selection of 027 isolates from each region. All 10 of the isolates from Quebec were of a single subtype, which was also dominant among isolates from Alberta (8/10 isolates) and British Columbia (6/8 isolates). Comparative sequencing of the tcdC repressor gene confirmed the documented 18-bp deletion and identified a second, single-base-pair deletion at position 117. Both deletions were conserved across all three provinces and were identified in a United Kingdom reference strain. The presence of a frameshift in the early portion of the tcdC gene implies serious functional disruption and may contribute to the hypervirulence of the 027 phenotype. PCR ribotype 027 strains appear to be widely distributed, to predate the Montreal outbreak, and to have measurable community presence in Western Canada.
Figures

References
-
- Alonso, R., A. Martin, T. Pelaez, M. Marin, M. Rodriguez-Creixems, and E. Bouza. 2005. An improved protocol for pulsed-field gel electrophoresis typing of Clostridium difficile. J. Med. Microbiol. 54:155-157. - PubMed
-
- Cohen, S. H., Y. J. Tang, and J. Silva, Jr. 2000. Analysis of the pathogenicity locus in Clostridium difficile strains. J. Infect. Dis. 181:659-663. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

