Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution
- PMID: 16760311
- PMCID: PMC1489541
- DOI: 10.1128/MMBR.00046-05
Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution
Abstract
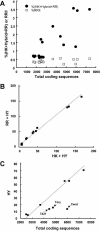
A survey of the already characterized and potential two-component protein sequences that exist in the nine complete and seven partially annotated cyanobacterial genome sequences available (as of May 2005) showed that the cyanobacteria possess a much larger repertoire of such proteins than most other bacteria. By analysis of the domain structure of the 1,171 potential histidine kinases, response regulators, and hybrid kinases, many various arrangements of about thirty different modules could be distinguished. The number of two-component proteins is related in part to genome size but also to the variety of physiological properties and ecophysiologies of the different strains. Groups of orthologues were defined, only a few of which have representatives with known physiological functions. Based on comparisons with the proposed phylogenetic relationships between the strains, the orthology groups show that (i) a few genes, some of them clustered on the genome, have been conserved by all species, suggesting their very ancient origin and an essential role for the corresponding proteins, and (ii) duplications, fusions, gene losses, insertions, and deletions, as well as domain shuffling, occurred during evolution, leading to the extant repertoire. These mechanisms are put in perspective with the different genetic properties that cyanobacteria have to achieve genome plasticity. This review is designed to serve as a basis for orienting further research aimed at defining the most ancient regulatory mechanisms and understanding how evolution worked to select and keep the most appropriate systems for cyanobacteria to develop in the quite different environments that they have successfully colonized.
Figures
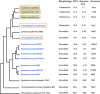
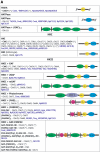
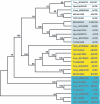
 ), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
 ), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
 ), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
 ), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
 ), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
 ), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
 ), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.
), protein that has no bacterial orthologue;
(+), putative sequencing error (see below). Putative proteins
that have no cyanobacterial orthologue are shown in blue and are
identified by their locus tag or GOI. Putative sequencing errors:
7120all1640-1639 (7A instead of 8A would create a frameshift, giving a
protein with 95% identity to Avar_400225220); the A of the
7421glr4211 stop codon can serve in the ATG initiation codon for
7421glr4212; Cwat_400841320 could form an HY with
Cwat_400841330; and the HATP-RR Cwat_400841330 could
form an HYI with
Cwat_400841320.


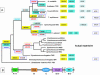
References
-
- Aiba, H., M. Nagaya, and T. Mizuno. 1993. Sensor and regulator proteins from the cyanobacterium Synechococcus sp. PCC7942 that belong to the bacterial signal-transduction protein families: implication in the adaptive response to phosphate limitation. Mol. Microbiol. 8:81-91. - PubMed
-
- Alves, R., and M. A. Savageau. 2003. Comparative analysis of prototype two-component systems with either bifunctional or monofunctional sensors: differences in molecular structure and physiological function. Mol. Microbiol. 48:25-51. - PubMed
-
- Anantharaman, V., and L. Aravind. 2000. Cache—a signaling domain common to animal Ca(2+)-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 25:535-537. - PubMed
-
- Anantharaman, V., E. V. Koonin, and L. Aravind. 2001. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 307:1271-1292. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

