Stable CpG hypomethylation of adipogenic promoters in freshly isolated, cultured, and differentiated mesenchymal stem cells from adipose tissue
- PMID: 16760426
- PMCID: PMC1525236
- DOI: 10.1091/mbc.e06-04-0322
Stable CpG hypomethylation of adipogenic promoters in freshly isolated, cultured, and differentiated mesenchymal stem cells from adipose tissue
Abstract
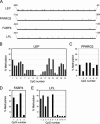
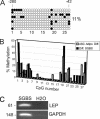
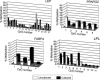
Mesenchymal stem cells from adipose tissue can differentiate into mesodermal lineages. Differentiation potential, however, varies between clones of adipose stem cells (ASCs), raising the hypothesis that epigenetic differences account for this variability. We report here a bisulfite sequencing analysis of CpG methylation of adipogenic (leptin [LEP], peroxisome proliferator-activated receptor gamma 2 [PPARG2], fatty acid-binding protein 4 [FABP4], and lipoprotein lipase [LPL]) promoters and of nonadipogenic (myogenin [MYOG], CD31, and GAPDH) loci in freshly isolated human ASCs and in cultured ASCs, in relation to gene expression and differentiation potential. Uncultured ASCs display hypomethylated adipogenic promoters, in contrast to myogenic and endothelial loci, which are methylated. Adipogenic promoters exhibit mosaic CpG methylation, on the basis of heterogeneous methylation between cells and of variation in the extent of methylation of a given CpG between donors, and both between and within clonal cell lines. DNA methylation reflects neither transcriptional status nor potential for gene expression upon differentiation. ASC culture preserves hypomethylation of adipogenic promoters; however, between- and within-clone mosaic methylation is detected. Adipogenic differentiation also maintains the overall CpG hypomethylation of LEP, PPARG2, FABP4, and LPL despite demethylation of specific CpGs and transcriptional induction. Furthermore, enhanced methylation at adipogenic loci in primary differentiated cells unrelated to adipogenesis argues for ASC specificity of the hypomethylated state of these loci. Therefore, mosaic hypomethylation of adipogenic promoters may constitute a molecular signature of ASCs, and DNA methylation does not seem to be a determinant of differentiation potential of these cells.
Figures






References
-
- Ayyanathan K., Lechner M. S., Bell P., Maul G. G., Schultz D. C., Yamada Y., Tanaka K., Torigoe K., Rauscher F. J., III, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. - PMC - PubMed
-
- Azuara V. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. - PubMed
-
- Ballas N., Grunseich C., Lu D. D., Speh J. C., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. - PubMed
-
- Bey L., Etienne J., Tse C., Brault D., Noe L., Raisonnier A., Arnault F., Hamilton M. T., Galibert F. Cloning, sequencing and structural analysis of 976 base pairs of the promoter sequence for the rat lipoprotein lipase gene. Comparison with the mouse and human sequences. Gene. 1998;209:31–38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

