Molecular anatomy of a speckle
- PMID: 16761280
- PMCID: PMC2563428
- DOI: 10.1002/ar.a.20336
Molecular anatomy of a speckle
Abstract
Direct localization of specific genes, RNAs, and proteins has allowed the dissection of individual nuclear speckles in relation to the molecular biology of gene expression. Nuclear speckles (aka SC35 domains) are essentially ubiquitous structures enriched for most pre-mRNA metabolic factors, yet their relationship to gene expression has been poorly understood. Analyses of specific genes and their spliced or mature mRNA strongly support that SC35 domains are hubs of activity, not stores of inert factors detached from gene expression. We propose that SC35 domains are hubs that spatially link expression of specific pre-mRNAs to rapid recycling of copious RNA metabolic complexes, thereby facilitating expression of many highly active genes. In addition to increasing the efficiency of each step, sequential steps in gene expression are structurally integrated at each SC35 domain, consistent with other evidence that the biochemical machineries for transcription, splicing, and mRNA export are coupled. Transcription and splicing are subcompartmentalized at the periphery, with largely spliced mRNA entering the domain prior to export. In addition, new findings presented here begin to illuminate the structural underpinnings of a speckle by defining specific perturbations of phosphorylation that promote disassembly or assembly of an SC35 domain in relation to other components. Results thus far are consistent with the SC35 spliceosome assembly factor as an integral structural component. Conditions that disperse SC35 also disperse poly(A) RNA, whereas the splicing factor ASF/SF2 can be dispersed under conditions in which SC35 or SRm300 remain as intact components of a core domain.
Figures
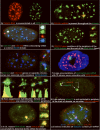
A) Poly(A) RNA (red) is concentrated in all SC35 domains (green).
B) Collagen 1A1 RNA (red) is consistently seen within an SC35 domain (green) (from: (Johnson et al., 2000))
C) Collagen 1A1 RNA (green) and collagen 1A2 RNA (red) can localize within a common SC35 (blue) (from: (Shopland et al., 2003))
D) Collagen 1A1 DNA hybridization (red) positions at the periphery of an SC35 domain (green).
E) Collagen 1A1 (green) and Col 1A2 (red) DNA hybridization shows multiple genes at the periphery of an SC35 domain (blue).
F) Dystrophin nuclear RNA accumulation (green) detected here using a cDNA probe to exons 1-11 (L. Kunkel Harvard School of Medicine, Boston, MA) is never observed to colocalize with SC35 domains (red) (Smith et al., 1999).
G) Exon suppression RNA hybridization with a labeled Collagen genomic probe (red) competed with excess cDNA shows that most introns are removed at the periphery of the SC35 domain (green) (from: (Johnson et al., 2000))
H) Comparison of collagen intron 24 and intron 26 shows that intron 24 is spliced later, and is not restricted to the periphery of domain or RNA track (green) (from: (Johnson et al., 2000))
I) In Osteogenesis Imperfecta, collagen mRNA from the mutant allele (lower signal and insets) retains intron 26 (green) throughout the mRNA track (red) and thus the Sc35 domain. The intron is removed from mRNA of the normal allele (upper signal) (from: (Johnson et al., 2000))
J) Visualization of three different mRNAs(beta actin, green, and collagen 1A1 and collagen 1A2, red) within Sc35 domains (blue), illustrates that half of the domains in this cell label with probes to just three genes. (from: Shopland, 2002])

References
-
- Barsh GS, Roush CL, Gelinas RE. DNA and Chromatin Structure of the Human alpha-1 (I) Collagen Gene. J.Biol.Chem. 1984;259(23):14906–14913. - PubMed
-
- Bisotto S, Lauriault P, Duval M, Vincent M. Colocalization of a high molecular mass phosphoprotein of the nuclear matrix (p255) with spliceosomes. J Cell Sci. 1995;108(Pt 5):1873–1882. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

