How plants tell the time
- PMID: 16761955
- PMCID: PMC1479754
- DOI: 10.1042/BJ20060484
How plants tell the time
Abstract
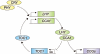
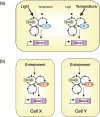
Plants, like all eukaryotes and most prokaryotes, have evolved sophisticated mechanisms for anticipating predictable environmental changes that arise due to the rotation of the Earth on its axis. These mechanisms are collectively termed the circadian clock. Many aspects of plant physiology, metabolism and development are under circadian control and a large proportion of the transcriptome exhibits circadian regulation. In the present review, we describe the advances in determining the molecular nature of the circadian oscillator and propose an architecture of several interlocking negative-feedback loops. The adaptive advantages of circadian control, with particular reference to the regulation of metabolism, are also considered. We review the evidence for the presence of multiple circadian oscillator types located in within individual cells and in different tissues.
Figures



References
-
- Pittendrigh C. S. Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 1993;55:17–54. - PubMed
-
- Barak S., Tobin E. M., Andronis C., Sugano S., Green R. M. All in good time: the Arabidopsis circadian clock. Trends Plant Sci. 2000;5:517–522. - PubMed
-
- Johnson C. H., Knight M. R., Kondo T., Masson P., Sedbook J., Haley A., Trewavas A. Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science. 1995;269:1863–1865. - PubMed
-
- McClung C. R., Salomé S. A., Michael T. P. The Arabidopsis circadian system. In: Somerville C. R., Meyerowitz E. M., editors. The Arabidopsis Book. Rockville: American Society of Plant Biologists; 2002. 10.1199/tab.0044, http://www.aspb.org/publications/arabidopsis/ - DOI - PMC - PubMed
-
- Harmer S. L., Panda S., Kay S. A. Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 2001;17:215–253. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

