Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing
- PMID: 16763550
- PMCID: PMC1500994
- DOI: 10.1038/sj.emboj.7601187
Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing
Abstract
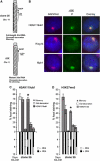
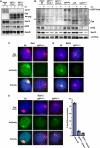
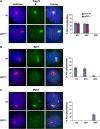
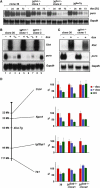
In mammals X inactivation is initiated by expression of Xist RNA and involves the recruitment of Polycomb repressive complex 1 (PRC1) and 2 (PRC2), which mediate chromosome-wide ubiquitination of histone H2A and methylation of histone H3, respectively. Here, we show that PRC1 recruitment by Xist RNA is independent of gene silencing. We find that Eed is required for the recruitment of the canonical PRC1 proteins Mph1 and Mph2 by Xist. However, functional Ring1b is recruited by Xist and mediates ubiquitination of histone H2A in Eed deficient embryonic stem (ES) cells, which lack histone H3 lysine 27 tri-methylation. Xist expression early in ES cell differentiation establishes a chromosomal memory, which allows efficient H2A ubiquitination in differentiated cells and is independent of silencing and PRC2. Our data show that Xist recruits PRC1 components by both PRC2 dependent and independent modes and in the absence of PRC2 function is sufficient for the establishment of Polycomb-based memory systems in X inactivation.
Figures

References
-
- Atsuta T, Fujimura S, Moriya H, Vidal M, Akasaka T, Koseki H (2001) Production of monoclonal antibodies against mammalian Ring1B proteins. Hybridoma 20: 43–46 - PubMed
-
- Birve A, Sengupta AK, Beuchle D, Larsson J, Kennison JA, Rasmuson-Lestander A, Muller J (2001) Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development 128: 3371–3379 - PubMed
-
- Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C et al. (1991) Characterization of a murine gene expressed from the inactive X chromosome. Nature 351: 325–329 - PubMed
-
- Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S (1991) Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351: 329–331 - PubMed
-
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF (1991a) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

