Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin
- PMID: 16763556
- PMCID: PMC1500862
- DOI: 10.1038/sj.emboj.7601178
Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin
Abstract
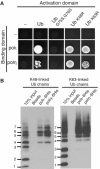
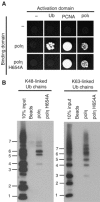
Y-family DNA polymerases have spacious active sites that can accommodate a wide variety of geometric distortions. As a consequence, they are considerably more error-prone than high-fidelity replicases. It is hardly surprising, therefore, that the in vivo activity of these polymerases is tightly regulated, so as to minimize their inadvertent access to primer-termini. We report here that one such mechanism employed by human cells relies on a specific and direct interaction between DNA polymerases iota and eta with ubiquitin (Ub). Indeed, we show that both polymerases interact noncovalently with free polyUb chains, as well as mono-ubiquitinated proliferating cell nuclear antigen (Ub-PCNA). Mutants of poliota (P692R) and poleta (H654A) were isolated that are defective in their interactions with polyUb and Ub-PCNA, whilst retaining their ability to interact with unmodified PCNA. Interestingly, the polymerase mutants exhibit significantly lower levels of replication foci in response to DNA damage, thereby highlighting the biological importance of the polymerase-Ub interaction in regulating the access of the TLS polymerases to stalled replication forks in vivo.
Figures




References
-
- Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M (2005) Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435: 979–982 - PubMed
-
- Bailly V, Lamb J, Sung P, Prakash S, Prakash L (1994) Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev 8: 811–820 - PubMed
-
- Bailly V, Lauder S, Prakash S, Prakash L (1997) Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem 272: 23360–23365 - PubMed
-
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I (2005) Novel ubiquitin-binding domains in Y-family polymerases regulate translesion DNA synthesis. Science 310: 1821–1824 - PubMed
-
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

