Roles for APIS and the 20S proteasome in adenovirus E1A-dependent transcription
- PMID: 16763564
- PMCID: PMC1500861
- DOI: 10.1038/sj.emboj.7601169
Roles for APIS and the 20S proteasome in adenovirus E1A-dependent transcription
Abstract
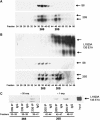
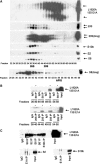
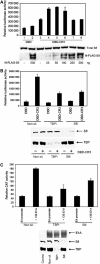
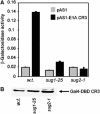
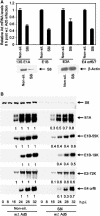
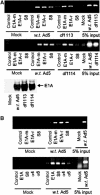
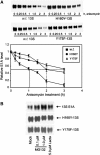
We have determined distinct roles for different proteasome complexes in adenovirus (Ad) E1A-dependent transcription. We show that the 19S ATPase, S8, as a component of 19S ATPase proteins independent of 20S (APIS), binds specifically to the E1A transactivation domain, conserved region 3 (CR3). Recruitment of APIS to CR3 enhances the ability of E1A to stimulate transcription from viral early gene promoters during Ad infection of human cells. The ability of CR3 to stimulate transcription in yeast is similarly dependent on the functional integrity of yeast APIS components, Sug1 and Sug2. The 20S proteasome is also recruited to CR3 independently of APIS and the 26S proteasome. Chromatin immunoprecipitation reveals that E1A, S8 and the 20S proteasome are recruited to both Ad early region gene promoters and early region gene sequences during Ad infection, suggesting their requirement in both transcriptional initiation and elongation. We also demonstrate that E1A CR3 transactivation and degradation sequences functionally overlap and that proteasome inhibitors repress E1A transcription. Taken together, these data demonstrate distinct roles for APIS and the 20S proteasome in E1A-dependent transactivation.
Figures



References
-
- Boyer TG, Berk AJ (1993) Functional interaction of adenovirus E1A with holo-TFIID. Genes Dev 7: 1810–1823 - PubMed
-
- Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ (1999) Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399: 276–279 - PubMed
-
- Chiang CM, Roeder RG (1995) Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 267: 531–536 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

