Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation
- PMID: 16763568
- PMCID: PMC1500856
- DOI: 10.1038/sj.emboj.7601159
Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation
Abstract
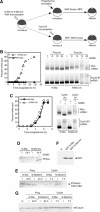
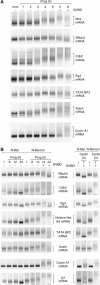
A strict temporal order of maternal mRNA translation is essential for meiotic cell cycle progression in oocytes of the frog Xenopus laevis. The molecular mechanisms controlling the ordered pattern of mRNA translational activation have not been elucidated. We report a novel role for the neural stem cell regulatory protein, Musashi, in controlling the translational activation of the mRNA encoding the Mos proto-oncogene during meiotic cell cycle progression. We demonstrate that Musashi interacts specifically with the polyadenylation response element in the 3' untranslated region of the Mos mRNA and that this interaction is necessary for early Mos mRNA translational activation. A dominant inhibitory form of Musashi blocks maternal mRNA cytoplasmic polyadenylation and meiotic cell cycle progression. Our data suggest that Musashi is a target of the initiating progesterone signaling pathway and reveal that late cytoplasmic polyadenylation element-directed mRNA translation requires early, Musashi-dependent mRNA translation. These findings indicate that Musashi function is necessary to establish the temporal order of maternal mRNA translation during Xenopus meiotic cell cycle progression.
Figures




References
-
- Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM (2006) The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21(WAF-1). Mol Cell Neurosci 31: 85–96 - PubMed
-
- Bernstein DS, Buter N, Stumpf C, Wickens M (2002) Analyzing mRNA–protein complexes using a yeast three-hybrid system. Methods 26: 123–141 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

