The intricate side of systems biology
- PMID: 16766654
- PMCID: PMC1480428
- DOI: 10.1073/pnas.0603337103
The intricate side of systems biology
Abstract
The combination of high-throughput methods of molecular biology with advanced mathematical and computational techniques has propelled the emergent field of systems biology into a position of prominence. Unthinkable a decade ago, it has become possible to screen and analyze the expression of entire genomes, simultaneously assess large numbers of proteins and their prevalence, and characterize in detail the metabolic state of a cell population. Although very important, the focus on comprehensive networks of biological components is only one side of systems biology. Complementing large-scale assessments, and sometimes at the risk of being forgotten, are more subtle analyses that rationalize the design and functioning of biological modules in exquisite detail. This intricate side of systems biology aims at identifying the specific roles of processes and signals in smaller, fully regulated systems by computing what would happen if these signals were lacking or organized in a different fashion. We exemplify this type of approach with a detailed analysis of the regulation of glucose utilization in Lactococcus lactis. This organism is exposed to alternating periods of glucose availability and starvation. During starvation, it accumulates an intermediate of glycolysis, which allows it to take up glucose immediately upon availability. This notable accumulation poses a nontrivial control task that is solved with an unusual, yet ingeniously designed and timed feedforward activation system. The elucidation of this control system required high-precision, dynamic in vivo metabolite data, combined with methods of nonlinear systems analysis, and may serve as a paradigm for multidisciplinary approaches to fine-scaled systems biology.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

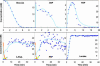

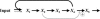

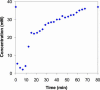
References
-
- Neves A. R., Ramos A., Nunes M. C., Kleerebezem M., Hugenholtz J., de Vos W. M., Almeida J., Santos H. Biotechnol. Bioeng. 1999;64:200–212. - PubMed
-
- Mason P. W., Carbone D. P., Cushman R. A., Waggoner A. S. J. Biol. Chem. 1981;256:1861–1866. - PubMed
-
- Voit E. O. Computational Analysis of Biochemical Systems. A Practical Guide for Biochemists and Molecular Biologists. Cambridge, U.K.: Cambridge Univ. Press; 2000.
-
- Neves A. R., Pool W. A., Kok J., Kuipers O. P., Santos H. FEMS Microbiol. Rev. 2005;29:531–554. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

