Circadian organization of the mammalian retina
- PMID: 16766660
- PMCID: PMC1480470
- DOI: 10.1073/pnas.0601940103
Circadian organization of the mammalian retina
Abstract
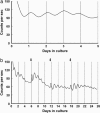
The mammalian retina contains an endogenous circadian pacemaker that broadly regulates retinal physiology and function, yet the cellular origin and organization of the mammalian retinal circadian clock remains unclear. Circadian clock neurons generate daily rhythms via cell-autonomous autoregulatory clock gene networks, and, thus, to localize circadian clock neurons within the mammalian retina, we have studied the cell type-specific expression of six core circadian clock genes in individual, identified mouse retinal neurons, as well as characterized the clock gene expression rhythms in photoreceptor degenerate rd mouse retinas. Individual photoreceptors, horizontal, bipolar, dopaminergic (DA) amacrines, catecholaminergic (CA) amacrines, and ganglion neurons were identified either by morphology or by a tyrosine hydroxylase (TH) promoter-driven red fluorescent protein (RFP) fluorescent reporter. Cells were collected, and their transcriptomes were subjected to multiplex single-cell RT-PCR for the core clock genes Period (Per) 1 and 2, Cryptochrome (Cry) 1 and 2, Clock, and Bmal1. Individual horizontal, bipolar, DA, CA, and ganglion neurons, but not photoreceptors, were found to coordinately express all six core clock genes, with the lowest proportion of putative clock cells in photoreceptors (0%) and the highest proportion in DA neurons (30%). In addition, clock gene rhythms were found to persist for >25 days in isolated, cultured rd mouse retinas in which photoreceptors had degenerated. Our results indicate that multiple types of retinal neurons are potential circadian clock neurons that express key elements of the circadian autoregulatory gene network and that the inner nuclear and ganglion cell layers of the mammalian retina contain functionally autonomous circadian clocks.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



References
-
- LaVail M. M. Science. 1976;194:1071–1074. - PubMed
-
- Terman J. S., Reme C. E., Terman M. Brain Res. 1993;605:256–264. - PubMed
-
- Grace M. S., Wang L. M., Pickard G. E., Besharse J. C., Menaker M. Brain Res. 1996;735:93–100. - PubMed
-
- Tosini G., Menaker M. Science. 1996;272:419–421. - PubMed
-
- Tosini G., Menaker M. Brain Res. 1998;789:221–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

