Membrane protein dynamics and detergent interactions within a crystal: a simulation study of OmpA
- PMID: 16766663
- PMCID: PMC1480439
- DOI: 10.1073/pnas.0600398103
Membrane protein dynamics and detergent interactions within a crystal: a simulation study of OmpA
Abstract
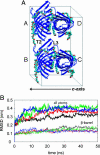
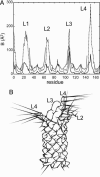
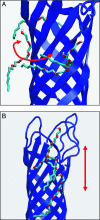
Molecular dynamics (MD) simulations are used to explore the dynamics of a membrane protein in its crystal environment. A 50-ns-duration simulation (at a temperature of 300 K) is performed for the crystallographic unit cell of the bacterial outer membrane protein OmpA. The unit cell contains four protein molecules, plus detergent molecules and water. An excellent correlation between simulated and experimental values of crystallographic B factors is observed. Effectively, 0.2 micros of protein trajectories are obtained, allowing a critical assessment of simulation quality. Some deficiency in conformational sampling is demonstrated, but averaging over multiple trajectories improves this limitation. The previously undescribed structure and dynamics of detergent molecules in a unit cell are reported here, providing insight into the interactions important in the formation and stabilization of the crystalline environment at room temperature. In particular, we show that at room temperature the detergent molecules form a dynamic, extended micellar structure spreading over adjacent OmpA monomers within the crystal.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

