The role of protein dynamics in thymidylate synthase catalysis: variants of conserved 2'-deoxyuridine 5'-monophosphate (dUMP)-binding Tyr-261
- PMID: 16768437
- PMCID: PMC2556892
- DOI: 10.1021/bi060152s
The role of protein dynamics in thymidylate synthase catalysis: variants of conserved 2'-deoxyuridine 5'-monophosphate (dUMP)-binding Tyr-261
Abstract
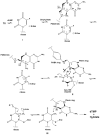
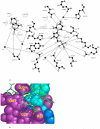
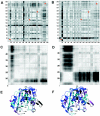
The enzyme thymidylate synthase (TS) catalyzes the reductive methylation of 2'-deoxyuridine 5'-monophosphate (dUMP) to 2'-deoxythymidine 5'-monophosphate. Using kinetic and X-ray crystallography experiments, we have examined the role of the highly conserved Tyr-261 in the catalytic mechanism of TS. While Tyr-261 is distant from the site of methyl transfer, mutants at this position show a marked decrease in enzymatic activity. Given that Tyr-261 forms a hydrogen bond with the dUMP 3'-O, we hypothesized that this interaction would be important for substrate binding, orientation, and specificity. Our results, surprisingly, show that Tyr-261 contributes little to these features of the mechanism of TS. However, the residue is part of the structural core of closed ternary complexes of TS, and conservation of the size and shape of the Tyr side chain is essential for maintaining wild-type values of kcat/Km. Moderate increases in Km values for both the substrate and cofactor upon mutation of Tyr-261 arise mainly from destabilization of the active conformation of a loop containing a dUMP-binding arginine. Besides binding dUMP, this loop has a key role in stabilizing the closed conformation of the enzyme and in shielding the active site from the bulk solvent during catalysis. Changes to atomic vibrations in crystals of a ternary complex of Escherichia coli Tyr261Trp are associated with a greater than 2000-fold drop in kcat/Km. These results underline the important contribution of dynamics to catalysis in TS.
Figures
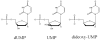



References
-
- Finer-Moore J, Fauman EB, Foster PG, Perry KM, Santi DV, Stroud RM. Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobacillus casei. J. Mol. Biol. 1993;232:1101–1116. - PubMed
-
- Climie S, Ruiz-Perez L, Gonzalez-Pacanowska D, Prapunwattana P, Cho SW, Stroud R, Santi DV. Saturation site-directed mutagenesis of thymidylate synthase. J. Biol. Chem. 1990;265:18776–9. - PubMed
-
- Climie SC, Carreras CW, Santi DV. A Complete Replacement Set of Amino Acids at the C-Terminus of Thymidylate Synthase: Quantitative Structure-Activity Relationships of Mutants of an Enzyme. Biochemistry. 1992;31:6032–6038. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

