Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT)
- PMID: 16769897
- PMCID: PMC1480433
- DOI: 10.1073/pnas.0601637103
Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT)
Abstract
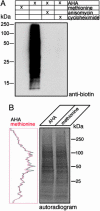
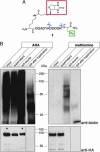
In both normal and pathological states, cells respond rapidly to environmental cues by synthesizing new proteins. The selective identification of a newly synthesized proteome has been hindered by the basic fact that all proteins, new and old, share the same pool of amino acids and thus are chemically indistinguishable. We describe here a technology, based on the cotranslational introduction of azide groups into proteins and the chemoselective tagging of azide-labeled proteins with an alkyne affinity tag, to separate and identify, specifically, the newly synthesized proteins in mammalian cells. Incorporation of the azide-bearing amino acid azidohomoalanine is unbiased, not toxic, and does not increase protein degradation. As a first demonstration of the method, we report the selective purification and identification of 195 metabolically labeled proteins with multidimensional liquid chromatography in-line with tandem MS. Furthermore, in combination with leucine-based mass tagging, candidates were immediately validated as newly synthesized proteins. The identified proteins, synthesized in a 2-h window, possess a broad range of biochemical properties and span most functional gene ontology categories. This technology makes it possible to address the temporal and spatial characteristics of newly synthesized proteomes in any cell type.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

References
-
- Lilley K. S., Friedman D. B. Exp. Rev. Proteomics. 2004;1:401–409. - PubMed
-
- Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. Nat. Biotechnol. 1999;17:994–999. - PubMed
-
- Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., et al. Mol. Cell Proteomics. 2004;3:1154–1169. - PubMed
-
- Washburn M. P., Ulaszek R., Deciu C., Schieltz D. M., Yates J. R., 3rd Anal. Chem. 2002;74:1650–1657. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

