Overexpression of SUR2A generates a cardiac phenotype resistant to ischemia
- PMID: 16770012
- PMCID: PMC2121651
- DOI: 10.1096/fj.05-5483com
Overexpression of SUR2A generates a cardiac phenotype resistant to ischemia
Abstract
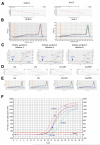
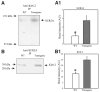
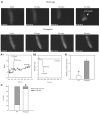
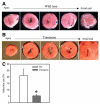
ATP-sensitive K+ (K(ATP)) channels are present in the sarcolemma of cardiac myocytes where they link membrane excitability with the cellular bioenergetic state. These channels are in vivo composed of Kir6.2, a pore-forming subunit, SUR2A, a regulatory subunit, and at least four accessory proteins. In the present study, real-time RT-PCR has demonstrated that of all six sarcolemmal K(ATP) channel-forming proteins, SUR2A was probably the least expressed protein. We have generated mice where the SUR2A was under the control of a cytomegalovirus promoter, a promoter that is more efficient than the native promoter. These mice had an increase in SUR2A mRNA/protein levels in the heart whereas levels of mRNAs of other channel-forming proteins were not affected at all. Imunoprecipitation/Western blot and patch clamp electrophysiology has shown an increase in K(ATP) channel numbers in the sarcolemma of transgenic mice. Cardiomyocytes from transgenic mice responded to hypoxia with shortening of action membrane potential and were significantly more resistant to this insult than cardiomyocytes from the wild-type. The size of myocardial infarction in response to ischemia-reperfusion was much smaller in hearts from transgenic mice compared to those in wild-type. We conclude that overexpression of SUR2A generates cardiac phenotype resistant to hypoxia/ischemia/reperfusion injury due at least in part to increase in levels of sarcolemmal K(ATP) channels.
Figures




References
-
- Kloner RA, Jennings RB. Consequences of brief ischaemia: stunning, preconditioning and their clinical implications: part 2. Circulation. 2001;104:3158–3167. - PubMed
-
- Kloner RA, Rezkalla SH. Cardiac protection during acute myocardial infarction: where do we stand in 2004? J. Am. Coll. Cardiol. 2004;44:276–286. - PubMed
-
- Noma A. ATP-regulated K+ channels in cardiac muscle. Nature. 1983;305:147–148. - PubMed
-
- Hanley PJ, Daut J. KATP channels and preconditioning: A re-examination of the role of mitochondrial KATP channels and an overview of alternative mechanisms. J. Mol. Cell. Cardiol. 2005;39:17–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- S18744/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 059528/Z/99/Z/JMW/CP/JF/WT_/Wellcome Trust/United Kingdom
- G0400608(71317)/MRC_/Medical Research Council/United Kingdom
- G0400608/MRC_/Medical Research Council/United Kingdom
- PG/02/091/14227/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

