Verification of predicted alternatively spliced Wnt genes reveals two new splice variants (CTNNB1 and LRP5) and altered Axin-1 expression during tumour progression
- PMID: 16772034
- PMCID: PMC1523213
- DOI: 10.1186/1471-2164-7-148
Verification of predicted alternatively spliced Wnt genes reveals two new splice variants (CTNNB1 and LRP5) and altered Axin-1 expression during tumour progression
Abstract
Background: Splicing processes might play a major role in carcinogenesis and tumour progression. The Wnt pathway is of crucial relevance for cancer progression. Therefore we focussed on the Wnt/beta-catenin signalling pathway in order to validate the expression of sequences predicted as alternatively spliced by bioinformatic methods. Splice variants of its key molecules were selected, which may be critical components for the understanding of colorectal tumour progression and may have the potential to act as biological markers. For some of the Wnt pathway genes the existence of splice variants was either proposed (e.g. beta-Catenin and CTNNB1) or described only in non-colon tissues (e.g. GSK3beta) or hitherto not published (e.g. LRP5).
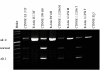
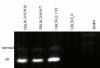
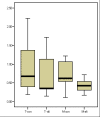
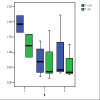
Results: Both splice variants--normal and alternative form--of all selected Wnt pathway components were found to be expressed in cell lines as well as in samples derived from tumour, normal and healthy tissues. All splice positions corresponded totally with the bioinformatical prediction as shown by sequencing. Two hitherto not described alternative splice forms (CTNNB1 and LRP5) were detected. Although the underlying EST data used for the bioinformatic analysis suggested a tumour-specific expression neither a qualitative nor a significant quantitative difference between the expression in tumour and healthy tissues was detected. Axin-1 expression was reduced in later stages and in samples from carcinomas forming distant metastases.
Conclusion: We were first to describe that splice forms of crucial genes of the Wnt-pathway are expressed in human colorectal tissue. Newly described splicefoms were found for beta-Catenin, LRP5, GSK3beta, Axin-1 and CtBP1. However, the predicted cancer specificity suggested by the origin of the underlying ESTs was neither qualitatively nor significant quantitatively confirmed. That let us to conclude that EST sequence data can give adequate hints for the existence of alternative splicing in tumour tissues. That no difference in the expression of these splice forms between cancerous tissues and normal mucosa was found, may indicate that the existence of different splice forms is of less significance for cancer formation as suggested by the available EST data. The currently available EST source is still insufficient to clearly deduce colon cancer specificity. More EST data from colon (tumour and healthy) is required to make reliable predictions.
Figures




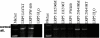


Similar articles
-
Identification of zinc-finger BED domain-containing 3 (Zbed3) as a novel Axin-interacting protein that activates Wnt/beta-catenin signaling.J Biol Chem. 2009 Mar 13;284(11):6683-9. doi: 10.1074/jbc.M807753200. Epub 2009 Jan 13. J Biol Chem. 2009. PMID: 19141611 Free PMC article.
-
Coordinated action of CK1 isoforms in canonical Wnt signaling.Mol Cell Biol. 2011 Jul;31(14):2877-88. doi: 10.1128/MCB.01466-10. Epub 2011 May 23. Mol Cell Biol. 2011. PMID: 21606194 Free PMC article.
-
Aberrant nuclear localization of beta-catenin without genetic alterations in beta-catenin or Axin genes in esophageal cancer.World J Surg Oncol. 2007 Feb 19;5:21. doi: 10.1186/1477-7819-5-21. World J Surg Oncol. 2007. PMID: 17309796 Free PMC article.
-
Molecular bases of the regulation of bone remodeling by the canonical Wnt signaling pathway.Curr Top Dev Biol. 2006;73:43-84. doi: 10.1016/S0070-2153(05)73002-7. Curr Top Dev Biol. 2006. PMID: 16782455 Review.
-
The many ways of Wnt in cancer.Curr Opin Genet Dev. 2007 Feb;17(1):45-51. doi: 10.1016/j.gde.2006.12.007. Curr Opin Genet Dev. 2007. PMID: 17208432 Review.
Cited by
-
Quantitative analysis of FJ 194940.1 gene expression in colon cancer and its association with clinicopathological parameters.Contemp Oncol (Pozn). 2013;17(1):45-50. doi: 10.5114/wo.2013.33773. Epub 2013 Mar 15. Contemp Oncol (Pozn). 2013. PMID: 23788961 Free PMC article.
-
Where Wnts went: the exploding field of Lrp5 and Lrp6 signaling in bone.J Bone Miner Res. 2009 Feb;24(2):171-8. doi: 10.1359/jbmr.081235. J Bone Miner Res. 2009. PMID: 19072724 Free PMC article. Review.
-
Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples.BMC Med Genomics. 2011 Jan 24;4:11. doi: 10.1186/1755-8794-4-11. BMC Med Genomics. 2011. PMID: 21261984 Free PMC article.
-
Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array.BMC Genomics. 2006 Dec 27;7:325. doi: 10.1186/1471-2164-7-325. BMC Genomics. 2006. PMID: 17192196 Free PMC article.
-
Harnessing low-density lipoprotein receptor protein 6 (LRP6) genetic variation and Wnt signaling for innovative diagnostics in complex diseases.Pharmacogenomics J. 2018 May 22;18(3):351-358. doi: 10.1038/tpj.2017.28. Epub 2017 Jul 11. Pharmacogenomics J. 2018. PMID: 28696417 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

