AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system
- PMID: 16774454
- PMCID: PMC1479700
- DOI: 10.1371/journal.pbio.0040229
AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system
Abstract
Activation of the insect innate immune system is dependent on a limited number of pattern recognition receptors (PRRs) capable of interacting with pathogen-associated molecular pattern. Here we report a novel role of an alternatively spliced hypervariable immunoglobulin domain-encoding gene, Dscam, in generating a broad range of PRRs implicated in immune defense in the malaria vector Anopheles gambiae. The mosquito Down syndrome cell adhesion molecule gene, AgDscam, has a complex genome organization with 101 exons that can produce over 31,000 potential alternative splice forms with different combinations of adhesive domains and interaction specificities. AgDscam responds to infection by producing pathogen challenge-specific splice form repertoires. Transient silencing of AgDscam compromises the mosquito's resistance to infections with bacteria and the malaria parasite Plasmodium. AgDscam is mediating phagocytosis of bacteria with which it can associate and defend against in a splice form-specific manner. AgDscam is a hypervariable PRR of the A. gambiae innate immune system.
Figures


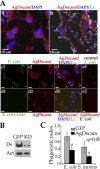

Comment in
-
A protean insect receptor holds the key to broad-based pathogen recognition.PLoS Biol. 2006 Jul;4(7):e246. doi: 10.1371/journal.pbio.0040246. Epub 2006 Jun 20. PLoS Biol. 2006. PMID: 20076613 Free PMC article. No abstract available.
References
-
- Hultmark D. Drosophila immunity: Paths and patterns . Curr Opin Immunol. 2003;15:12–19. - PubMed
-
- Meister M. Blood cells of Drosophila Cell lineages and role in host defense . Curr Opin Immunol. 2004;16:10–15. - PubMed
-
- Dimopoulos G. Insect immunity and its implication in mosquito-malaria interactions. Cell Microbiol. 2003;5:3–14. - PubMed
-
- Hoffmann JA. The immune response of Drosophila . Nature. 2003;426:33–38. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

