Ribosomal protein S1 promotes transcriptional cycling
- PMID: 16775305
- PMCID: PMC1524893
- DOI: 10.1261/rna.2321606
Ribosomal protein S1 promotes transcriptional cycling
Abstract
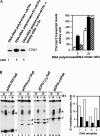
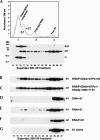
Prokaryotic RNA polymerases are capable of efficient, continuous synthesis of RNA in vivo, yet purified polymerase-DNA model systems for RNA synthesis typically produce only a limited number of catalytic turnovers. Here, we report that the ribosomal protein S1--which plays critical roles in translation initiation and elongation in Escherichia coli and is believed to stabilize mRNA on the ribosome--is a potent activator of transcriptional cycling in vitro. Deletion of the two C-terminal RNA-binding modules--out of a total of six loosely homologous RNA-binding modules present in S1--resulted in a near-loss of the ability of S1 to enhance transcription, whereas disruption of the very last C-terminal RNA-binding module had only a mild effect. We propose that, in vivo, cooperative interaction of multiple RNA-binding modules in S1 may enhance the transcript release from RNA polymerase, alleviating its inhibitory effect and enabling the core enzyme for continuous reinitiation of transcription.
Figures





References
-
- Arndt K.M., Chamberlin M.J. Transcription termination in Escherichia coli. Measurement of the rate of enzyme release from Rho-independent terminators. J. Mol. Biol. 1988;202:271–285. - PubMed
-
- Berlin V., Yanofsky C. Release of transcript and template during transcription termination at the trp operon attenuator. J.Biol. Chem. 1983;258:1714–1719. - PubMed
-
- Bycroft M., Hubbard T.J.P., Proctor M., Freund S.M.V., Murzin A.G. The solution structure of the S1 RNA binding domain: A member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
