Cloning, expression, and functional characterization of the equine herpesvirus 1 DNA polymerase and its accessory subunit
- PMID: 16775312
- PMCID: PMC1488933
- DOI: 10.1128/JVI.02551-05
Cloning, expression, and functional characterization of the equine herpesvirus 1 DNA polymerase and its accessory subunit
Abstract
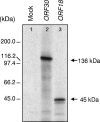
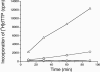
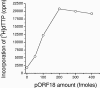
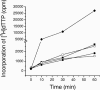
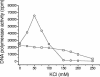
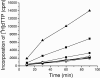
We report the expression and characterization of the putative catalytic subunit (pORF30) and accessory protein (pORF18) of equine herpesvirus 1 DNA polymerase, which are encoded by open reading frames 30 and 18 and are homologous to herpes simplex virus type 1 UL30 and UL42, respectively. In vitro transcription-translation of open reading frames 30 and 18 generated proteins of 136 and 45 kDa, respectively. In vitro-expressed pORF30 possessed basal DNA polymerase activity that was stimulated by pORF18, as measured by DNA polymerase assays in vitro. Purified baculovirus-expressed pORF30 exhibited DNA polymerase activity similar to that of the in vitro-expressed protein, and baculovirus-expressed pORF18 could stimulate both nucleotide incorporation and long-chain DNA synthesis by pORF30 in a dose- and time-dependent manner. The salt optima for activity of both pORF30 and the holoenzyme were substantially different from those for other herpesvirus DNA polymerases. As demonstrated by yeast two-hybrid assays, pORF30 and pORF18 could physically interact, most likely with a 1:1 stoichiometry. Finally, by mutational analysis of the 1,220-residue pORF30, we demonstrated that the extreme C terminus of pORF30 is important for physical and functional interaction with the accessory protein, as reported for UL30 and other herpesvirus DNA polymerases. In addition, a C-proximal region of pORF30, corresponding to residues 1114 to 1172, is involved in binding to, and stimulation by, pORF18. Taken together, the results indicate that pORF30 and pORF18 are the equine herpesvirus 1 counterparts of herpes simplex virus type 1 UL30 and UL42 and share many, but not all, of their characteristics.
Figures


Similar articles
-
The catalytic subunit of herpes simplex virus type 1 DNA polymerase contains a nuclear localization signal in the UL42-binding region.Virology. 2000 Jul 20;273(1):139-48. doi: 10.1006/viro.2000.0390. Virology. 2000. PMID: 10891416
-
Analysis of in vitro activities of herpes simplex virus type 1 UL42 mutant proteins: correlation with in vivo function.Virology. 2000 Sep 30;275(2):373-90. doi: 10.1006/viro.2000.0506. Virology. 2000. PMID: 10998337
-
DNA and protein interactions of the small subunit of herpes simplex virus type 1 DNA polymerase.Virology. 1999 Jan 5;253(1):55-64. doi: 10.1006/viro.1998.9491. Virology. 1999. PMID: 9887318
-
Disruption of the interactions between the subunits of herpesvirus DNA polymerases as a novel antiviral strategy.Clin Microbiol Infect. 2005 Jun;11(6):437-46. doi: 10.1111/j.1469-0691.2005.01149.x. Clin Microbiol Infect. 2005. PMID: 15882193 Review.
-
The baculovirus 10-kDa protein.J Invertebr Pathol. 1997 Jul;70(1):1-17. doi: 10.1006/jipa.1997.4675. J Invertebr Pathol. 1997. PMID: 9217463 Review. No abstract available.
Cited by
-
The early UL31 gene of equine herpesvirus 1 encodes a single-stranded DNA-binding protein that has a nuclear localization signal sequence at the C-terminus.Virology. 2012 Oct 25;432(2):306-15. doi: 10.1016/j.virol.2012.05.031. Epub 2012 Jun 20. Virology. 2012. PMID: 22721961 Free PMC article.
-
A point mutation in a herpesvirus polymerase determines neuropathogenicity.PLoS Pathog. 2007 Nov;3(11):e160. doi: 10.1371/journal.ppat.0030160. PLoS Pathog. 2007. PMID: 17997600 Free PMC article.
-
Identification of conserved amino acids in the herpes simplex virus type 1 UL8 protein required for DNA synthesis and UL52 primase interaction in the virus replisome.J Biol Chem. 2012 Sep 28;287(40):33142-52. doi: 10.1074/jbc.M112.356782. Epub 2012 Jul 30. J Biol Chem. 2012. PMID: 22851167 Free PMC article.
-
Regulated transport into the nucleus of herpesviridae DNA replication core proteins.Viruses. 2013 Sep 16;5(9):2210-34. doi: 10.3390/v5092210. Viruses. 2013. PMID: 24064794 Free PMC article.
References
-
- Allen, G. P., and J. T. Bryans. 1986. Molecular epizootiology, pathogenesis, and prophylaxis of equine herpesvirus-1 infections. Prog. Vet. Microbiol. Immunol. 2:78-144. - PubMed
-
- Appleton, B. A., A. Loregian, D. J. Filman, D. M. Coen, and J. M. Hogle. 2004. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol. Cell 15:233-244. - PubMed
-
- Bishop, D. H. 1992. Baculovirus expression vectors. Semin. Virol. 3:253-264.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

