Cumulative mutations of ubiquitin acceptor sites in human immunodeficiency virus type 1 gag cause a late budding defect
- PMID: 16775314
- PMCID: PMC1488962
- DOI: 10.1128/JVI.02177-05
Cumulative mutations of ubiquitin acceptor sites in human immunodeficiency virus type 1 gag cause a late budding defect
Abstract
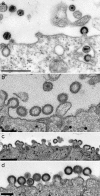
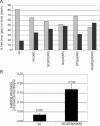
The p6 domain of human immunodeficiency virus type 1 (HIV-1) Gag has long been known to be monoubiquitinated. We have previously shown that the MA, CA, and NC domains are also monoubiquitinated at low levels (E. Gottwein and H. G. Krausslich, J. Virol. 79:9134-9144, 2005). While several lines of evidence support a role for ubiquitin in virus release, the relevance of Gag ubiquitination is unclear. To directly address the function of Gag ubiquitination, we constructed Gag variants in which lysine residues in the NC, SP2, and p6 domains were mutated to arginine either in individual domains or in combination. Using these mutants, we showed that in addition to MA, CA, NC, and p6, SP2 is also mono- or di-ubiquitinated at levels comparable to those of the other domains. Replacement of all lysine residues in only one of the domains had minor effects on virus release, while cumulative mutations in NC and SP2 or in NC and p6 resulted in an accumulation of late budding structures, as observed by electron microscopy analysis. Strikingly, replacement of all lysine residues downstream of CA led to a significant reduction in virus release kinetics and a fivefold accumulation of late viral budding structures compared to wild-type levels. These results indicate that ubiquitination of lysine residues in Gag in the vicinity of the viral late domain is important for HIV-1 budding, while no specific lysine residue may be needed and individual domains can functionally substitute. This is consistent with Gag ubiquitination being functionally involved in a transient protein interaction network at the virus budding site.
Figures





Similar articles
-
Analysis of human immunodeficiency virus type 1 Gag ubiquitination.J Virol. 2005 Jul;79(14):9134-44. doi: 10.1128/JVI.79.14.9134-9144.2005. J Virol. 2005. PMID: 15994808 Free PMC article.
-
Ubiquitination of HIV-1 and MuLV Gag.Virology. 2000 Dec 5;278(1):111-21. doi: 10.1006/viro.2000.0648. Virology. 2000. PMID: 11112487
-
Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function.Proc Natl Acad Sci U S A. 2002 Jan 22;99(2):955-60. doi: 10.1073/pnas.032511899. Proc Natl Acad Sci U S A. 2002. PMID: 11805336 Free PMC article.
-
The role of viral and cellular proteins in the budding of human immunodeficiency virus.Acta Virol. 2006;50(2):75-85. Acta Virol. 2006. PMID: 16808324 Review.
-
HIV-1 matrix protein: a mysterious regulator of the viral life cycle.Virus Res. 2007 Mar;124(1-2):1-11. doi: 10.1016/j.virusres.2006.07.001. Epub 2007 Jan 8. Virus Res. 2007. PMID: 17210199 Review.
Cited by
-
The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process.J Virol. 2010 May;84(9):4725-36. doi: 10.1128/JVI.02478-09. Epub 2010 Feb 17. J Virol. 2010. PMID: 20164219 Free PMC article.
-
Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding.J Virol. 2011 Apr;85(7):3546-56. doi: 10.1128/JVI.02045-10. Epub 2010 Dec 29. J Virol. 2011. PMID: 21191027 Free PMC article.
-
Viral takeover of the host ubiquitin system.Front Microbiol. 2011 Jul 28;2:161. doi: 10.3389/fmicb.2011.00161. eCollection 2011. Front Microbiol. 2011. PMID: 21847386 Free PMC article.
-
PJA2 ubiquitinates the HIV-1 Tat protein with atypical chain linkages to activate viral transcription.Sci Rep. 2017 Mar 27;7:45394. doi: 10.1038/srep45394. Sci Rep. 2017. PMID: 28345603 Free PMC article.
-
HIV-1 Gag release from yeast reveals ESCRT interaction with the Gag N-terminal protein region.J Biol Chem. 2020 Dec 25;295(52):17950-17972. doi: 10.1074/jbc.RA120.014710. Epub 2020 Sep 28. J Biol Chem. 2020. PMID: 32994219 Free PMC article.
References
-
- Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. - PubMed
-
- Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

