Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells
- PMID: 16775325
- PMCID: PMC1488946
- DOI: 10.1128/JVI.00211-06
Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells
Abstract
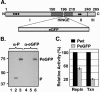
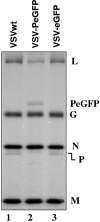
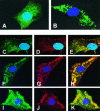
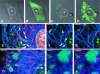
The phosphoprotein (P) of vesicular stomatitis virus (VSV) is a subunit of the viral RNA polymerase. In previous studies, we demonstrated that insertion of 19 amino acids in the hinge region of the protein had no significant effect on P protein function. In the present study, we inserted full-length enhanced green fluorescent protein (eGFP) in frame into the hinge region of P and show that the fusion protein (PeGFP) is functional in viral genome transcription and replication, albeit with reduced activity. A recombinant vesicular stomatitis virus encoding PeGFP in place of the P protein (VSV-PeGFP), which possessed reduced growth kinetics compared to the wild-type VSV, was recovered. Using the recombinant VSV-PeGFP, we show that the viral replication proteins and the de novo-synthesized RNA colocalize to sites throughout the cytoplasm, indicating that replication and transcription are not confined to any particular region of the cytoplasm. Real-time imaging of the cells infected with the eGFP-tagged virus revealed that, following synthesis, the nucleocapsids are transported toward the cell periphery via a microtubule (MT)-mediated process, and the nucleocapsids were seen to be closely associated with mitochondria. Treatment of cells with nocodazole or Colcemid, drugs known to inhibit MT polymerization, resulted in accumulation of the nucleocapsids around the nucleus and also led to inhibition of infectious-virus production. These findings are compatible with a model in which the progeny viral nucleocapsids are transported toward the cell periphery by MT and the transport may be facilitated by mitochondria.
Figures



References
-
- Canter, D. M., and J. Perrault. 1996. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding. Virology 219:376-386. - PubMed
-
- Charpilienne, A., M. Nejmeddine, M. Berois, N. Parez, E. Neumann, E. Hewat, G. Trugnan, and J. Cohen. 2001. Individual rotavirus-like particles containing 120 molecules of fluorescent protein are visible in living cells. J. Biol. Chem. 276:29361-29367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

