A Tlr7 translocation accelerates systemic autoimmunity in murine lupus
- PMID: 16777955
- PMCID: PMC1502563
- DOI: 10.1073/pnas.0603912103
A Tlr7 translocation accelerates systemic autoimmunity in murine lupus
Abstract
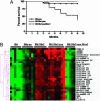
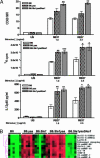
The y-linked autoimmune accelerating (yaa) locus is a potent autoimmune disease allele. Transcription profiling of yaa-bearing B cells revealed the overexpression of a cluster of X-linked genes that included Tlr7. FISH analysis demonstrated the translocation of this segment onto the yaa chromosome. The resulting overexpression of Tlr7 increased in vitro responses to Toll-like receptor (TLR) 7 signaling in all yaa-bearing males. B6.yaa mice are not overtly autoimmune, but the addition of Sle1, which contains the autoimmune-predisposing Slam/Cd2 haplotype, causes the development of fatal lupus with numerous immunological aberrations. B6.Sle1yaa CD4 T cells develop the molecular signature for T(FH) cells and also show expression changes in numerous cytokines and chemokines. Disease development and all component autoimmune phenotypes were inhibited by Sles1, a potent suppressor locus. Sles1 had no effect on yaa-enhanced TLR7 signaling in vitro, and these data place Sles1 downstream from the lesion in innate immune responses mediated by TLR7, suggesting that Sles1 modulates the activation of adaptive immunity in response to innate immune signaling.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
-
- Wakeland E. K., Liu K., Graham R. R., Behrens T. W. Immunity. 2001;15:397–408. - PubMed
-
- Wandstrat A. E., Wakeland E. K. Nat. Immunol. 2001;2:802–809. - PubMed
-
- Hudgins C. C., Steinberg R. T., Klinman D. M., Reeves M. J., Steinberg A. D. J. Immunol. 1985;134:3849–3854. - PubMed
-
- Liu K., Liang C., Liang Z., Tus K., Wakeland E. K. J. Immunol. 2005;174:1630–1637. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

