The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement
- PMID: 16777964
- PMCID: PMC1502546
- DOI: 10.1073/pnas.0602249103
The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement
Abstract
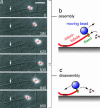
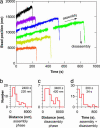
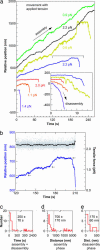
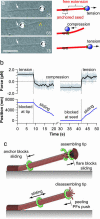
Kinetochores remain attached to microtubule (MT) tips during mitosis even as the tips assemble and disassemble under their grip, allowing filament dynamics to produce force and move chromosomes. The specific proteins that mediate tip attachment are uncertain, and the mechanism of MT-dependent force production is unknown. Recent work suggests that the Dam1 complex, an essential component of kinetochores in yeast, may contribute directly to kinetochore-MT attachment and force production, perhaps by forming a sliding ring encircling the MT. To test these hypotheses, we developed an in vitro motility assay where beads coated with pure recombinant Dam1 complex were bound to the tips of individual dynamic MTs. The Dam1-coated beads remained tip-bound and underwent assembly- and disassembly-driven movement over approximately 3 microm, comparable to chromosome displacements in vivo. Dam1-based attachments to assembling tips were robust, supporting 0.5-3 pN of tension applied with a feedback-controlled optical trap as the MTs lengthened approximately 1 microm. The attachments also harnessed energy from MT disassembly to generate movement against tension. Reversing the direction of force (i.e., switching to compressive force) caused the attachments to disengage the tip and slide over the filament, but sliding was blocked by areas where the MT was anchored to a coverslip, consistent with a coupling structure encircling the filament. Our findings demonstrate how the Dam1 complex may contribute directly to MT-driven chromosome movement.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

