Regulation of transcription factor latency by receptor-activated proteolysis
- PMID: 16778074
- PMCID: PMC1482476
- DOI: 10.1101/gad.374206
Regulation of transcription factor latency by receptor-activated proteolysis
Abstract
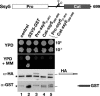
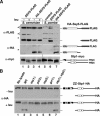
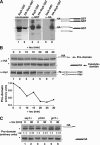
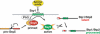
The transcription factor Stp1 is endoproteolytically processed in response to extracellular amino acids by the plasma membrane SPS (Ssy1-Ptr3-Ssy5)-sensor. Processed Stp1, lacking a cytoplasmic retention motif, enters the nucleus and induces amino acid transporter gene expression. The SPS-sensor component Ssy5 is a chymotrypsin-like protease with a Pro-domain and a catalytic domain. The Pro-domain, required for protease maturation, is autolytically cleaved from the catalytic domain but remains associated, forming an inactive protease complex that binds Stp1. Stp1 is processed only after amino acid-induced signals cause the dissociation of the inhibitory Pro-domain. Our findings demonstrate that gene expression can be controlled by regulating the enzymatic activity of an intracellular endoprotease.
Figures

References
-
- Andréasson C. Ph.D. thesis, Karolinska Institute. Karolinska Institute Press; Stockholm, Sweden: 2004. “Ligand-activated proteolysis in nutrient signaling.”.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
