Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint
- PMID: 16782009
- PMCID: PMC2749311
- DOI: 10.1016/j.cub.2006.04.043
Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint
Abstract
In the presence of unattached/weakly attached kinetochores, the spindle assembly checkpoint (SAC) delays exit from mitosis by preventing the anaphase-promoting complex (APC)-mediated proteolysis of cyclin B, a regulatory subunit of cyclin-dependent kinase 1 (Cdk1). Like all checkpoints, the SAC does not arrest cells permanently, and escape from mitosis in the presence of an unsatisfied SAC requires that cyclin B/Cdk1 activity be inhibited. In yeast , and likely Drosophila, this occurs through an "adaptation" process involving an inhibitory phosphorylation on Cdk1 and/or activation of a cyclin-dependent kinase inhibitor (Cdki). The mechanism that allows vertebrate cells to escape mitosis when the SAC cannot be satisfied is unknown. To explore this issue, we conducted fluorescence microscopy studies on rat kangaroo (PtK) and human (RPE1) cells dividing in the presence of nocodazole. We find that in the absence of microtubules (MTs), escape from mitosis occurs in the presence of an active SAC and requires cyclin B destruction. We also find that cyclin B is progressively destroyed during the block by a proteasome-dependent mechanism. Thus, vertebrate cells do not adapt to the SAC. Rather, our data suggest that in normal cells, the SAC cannot prevent a slow but continuous degradation of cyclin B that ultimately drives the cell out of mitosis.
Figures
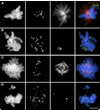
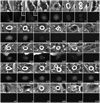


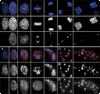
References
-
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr. Opin. Cell Biol. 1996;8:773–780. - PubMed
-
- Minshull J, Straight A, Rudner AD, Dernburg AF, Belmont A, Murray AW. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 1996;6:1609–1620. - PubMed
-
- Novak B, Toth A, Csikasz-Nagy A, Gyorffy B, Tyson JJ, Nasmyth K. Finishing the cell cycle. J. Theor. Biol. 1999;199:223–233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

