Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment
- PMID: 16782035
- PMCID: PMC1626529
- DOI: 10.1016/j.immuni.2006.04.008
Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment
Abstract
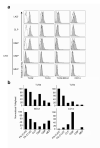
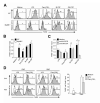
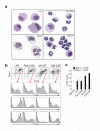
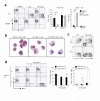
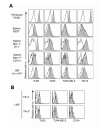
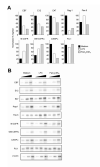
Toll-like receptors (TLRs) are best known for their ability to recognize microbial or viral components and initiate innate immune responses. We showed here that TLRs and their coreceptors were expressed by multipotential hematopoietic stem cells, whose cell cycle entry was triggered by TLR ligation. TLR expression also extended to some of the early hematopoietic progenitors, although not the progenitor cells dedicated to megakaryocyte and erythroid differentiation. TLR signaling via the Myd88 adaptor protein drove differentiation of myeloid progenitors, bypassing some normal growth and differentiation requirements, and also drove lymphoid progenitors to become dendritic cells. CD14 contributed to the efficiency of lipopolysaccharide (LPS) recognition by stem and progenitor cells, and LPS interacted directly with the TLR4/MD-2 complex on these cells in bone marrow. Thus, the preferential pathogen-mediated stimulation of myeloid differentiation pathways may provide a means for rapid replenishment of the innate immune system during infection.
Figures










Comment in
-
Outside influence: TLRs direct hematopoietic cell fates.Immunity. 2006 Jun;24(6):667-669. doi: 10.1016/j.immuni.2006.06.007. Immunity. 2006. PMID: 16782021 Review.
References
-
- Akashi K, Traver D, Miyamoto M, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. - PubMed
-
- Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 2003;3:169–176. - PubMed
-
- Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304:1014–1018. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

