Potential roles for ubiquitin and the proteasome during ribosome biogenesis
- PMID: 16782897
- PMCID: PMC1489179
- DOI: 10.1128/MCB.02227-05
Potential roles for ubiquitin and the proteasome during ribosome biogenesis
Erratum in
- Mol Cell Biol. 2006 Aug;26(16):6308
Abstract
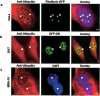
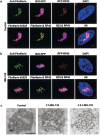
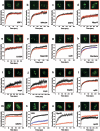
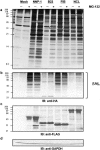
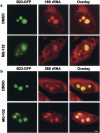
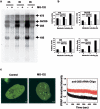
We have investigated the possible involvement of the ubiquitin-proteasome system (UPS) in ribosome biogenesis. We find by immunofluorescence that ubiquitin is present within nucleoli and also demonstrate by immunoprecipitation that complexes associated with pre-rRNA processing factors are ubiquitinated. Using short proteasome inhibition treatments, we show by fluorescence microscopy that nucleolar morphology is disrupted for some but not all factors involved in ribosome biogenesis. Interference with proteasome degradation also induces the accumulation of 90S preribosomes, alters the dynamic properties of a number of processing factors, slows the release of mature rRNA from the nucleolus, and leads to the depletion of 18S and 28S rRNAs. Together, these results suggest that the UPS is probably involved at many steps during ribosome biogenesis, including the maturation of the 90S preribosome.
Figures


References
-
- Andersen, J. S., Y. W. Lam, A. K. Leung, S. E. Ong, C. E. Lyon, A. I. Lamond, and M. Mann. 2005. Nucleolar proteome dynamics. Nature 433:77-83. - PubMed
-
- Arabi, A., C. Rustum, E. Hallberg, and A. P. Wright. 2003. Accumulation of c-Myc and proteasomes at the nucleoli of cells containing elevated c-Myc protein levels. J. Cell Sci. 116:1707-1717. - PubMed
-
- Boulanger, J., A. Vezina, S. Mongrain, F. Boudreau, N. Perreault, B. A. Auclair, J. Laine, C. Asselin, and N. Rivard. 2005. Cdk2-dependent phosphorylation of homeobox transcription factor CDX2 regulates its nuclear translocation and proteasome-mediated degradation in human intestinal epithelial cells. J. Biol. Chem. 280:18095-18107. - PubMed
-
- Carmo-Fonseca, M., L. Mendes-Soares, and I. Campos. 2000. To be or not to be in the nucleolus. Nat. Cell Biol. 2:E107-E112. - PubMed
-
- Chan, Y. L., K. Suzuki, and I. G. Wool. 1995. The carboxyl extensions of two rat ubiquitin fusion proteins are ribosomal proteins S27a and L40. Biochem. Biophys. Res. Commun. 215:682-690. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
