Poor performance of bootstrap confidence intervals for the location of a quantitative trait locus
- PMID: 16783000
- PMCID: PMC1569776
- DOI: 10.1534/genetics.106.061549
Poor performance of bootstrap confidence intervals for the location of a quantitative trait locus
Abstract
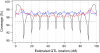
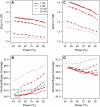
The aim of many genetic studies is to locate the genomic regions (called quantitative trait loci, QTL) that contribute to variation in a quantitative trait (such as body weight). Confidence intervals for the locations of QTL are particularly important for the design of further experiments to identify the gene or genes responsible for the effect. Likelihood support intervals are the most widely used method to obtain confidence intervals for QTL location, but the nonparametric bootstrap has also been recommended. Through extensive computer simulation, we show that bootstrap confidence intervals behave poorly and so should not be used in this context. The profile likelihood (or LOD curve) for QTL location has a tendency to peak at genetic markers, and so the distribution of the maximum-likelihood estimate (MLE) of QTL location has the unusual feature of point masses at genetic markers; this contributes to the poor behavior of the bootstrap. Likelihood support intervals and approximate Bayes credible intervals, on the other hand, are shown to behave appropriately.
Figures




References
-
- Beran, R., 2003. The impact of the bootstrap on statistical algorithms and theory. Stat. Sci. 18: 175–184.
-
- Broman, K. W., H. Wu, Ś. Sen and G. A. Churchill, 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. - PubMed
-
- Dempster, A. P., N. M. Laird and D. B. Rubin, 1977. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. B 39: 1–38.
-
- Haley, C. S., and S. A. Knott, 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

