From genetical genomics to systems genetics: potential applications in quantitative genomics and animal breeding
- PMID: 16783637
- PMCID: PMC3906707
- DOI: 10.1007/s00335-005-0169-x
From genetical genomics to systems genetics: potential applications in quantitative genomics and animal breeding
Abstract
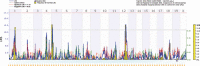
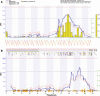
This article reviews methods of integration of transcriptomics (and equally proteomics and metabolomics), genetics, and genomics in the form of systems genetics into existing genome analyses and their potential use in animal breeding and quantitative genomic modeling of complex traits. Genetical genomics or the expression quantitative trait loci (eQTL) mapping method and key findings in this research are reviewed. Various procedures and potential uses of eQTL mapping, global linkage clustering, and systems genetics are illustrated using actual analysis on recombinant inbred lines of mice with data on gene expression (for diabetes- and obesity-related genes), pathway, and single nucleotide polymorphism (SNP) linkage maps. Experimental and bioinformatics difficulties and possible solutions are discussed. The main uses of this systems genetics approach in quantitative genomics were shown to be in refinement of the identified QTL, candidate gene and SNP discovery, understanding gene-environment and gene-gene interactions, detection of candidate regulator genes/eQTL, discriminating multiple QTL/eQTL, and detection of pleiotropic QTL/eQTL, in addition to its use in reconstructing regulatory networks. The potential uses in animal breeding are direct selection on heritable gene expression measures, termed "expression assisted selection," and genetical genomic selection of both QTL and eQTL based on breeding values of the respective genes, termed "expression-assisted evaluation."
Figures



References
-
- Bing N, Hoeschele I, Ye KY, Eilertsen KJ. Finite mixture model analysis of microarray expression data on samples of uncertain biological type with application to reproductive efficiency. Vet Immunol Immunopathol. 2005;105:187–196. - PubMed
-
- Brazhnik P, de la Fuente A, Mendes P. Gene networks: how to put the function in genomics. Trends Biotechnol. 2002;20:467–472. - PubMed
-
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. - PubMed
-
- Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using “genetical genomics.”. Nat Genet. 2005;37:225–232. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

