Activation of the global gene regulator PrrA (RegA) from Rhodobacter sphaeroides
- PMID: 16784239
- PMCID: PMC2517121
- DOI: 10.1021/bi060683g
Activation of the global gene regulator PrrA (RegA) from Rhodobacter sphaeroides
Abstract
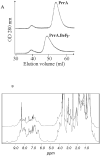
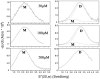
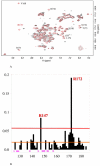
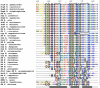
PrrA is a global transcription regulator activated upon phosphorylation by its cognate kinase PrrB in response to low oxygen levels in Rhodobacter sphaeroides. Here we show by gel filtration, analytical ultracentrifugation, and NMR diffusion measurements that treatment of PrrA with a phosphate analogue, BeF(3)(-), results in dimerization of the protein, producing a protein that binds DNA. No dimeric species was observed in the absence of BeF(3)(-). Upon addition of BeF(3)(-), the inhibitory activity of the N-terminal domain on the C-terminal DNA-binding domain is relieved, after which PrrA becomes capable of binding DNA as a dimer. The interaction surface of the DNA-binding domain with the regulatory domain of PrrA is identified by NMR as being a well-conserved region centered on helix alpha6, which is on the face opposite from the DNA recognition helix. This suggests that there is no direct blockage of DNA binding in the inactive state but rather that PrrA dimerization promotes a correct arrangement of two adjacent DNA-binding domains that recognizes specific DNA binding sequences.
Figures


References
-
- Swem LR, Elsen S, Bird TH, Swem DL, Koch HG, Myllykallio H, Daldal F, Bauer CE. The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J. Mol. Biol. 2001;309:121–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

