The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells
- PMID: 16785433
- PMCID: PMC1502513
- DOI: 10.1073/pnas.0603729103
The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells
Abstract
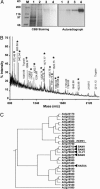
AtPep1 is a 23-aa endogenous peptide elicitor from Arabidopsis leaves that signals the activation of components of the innate immune response against pathogens. Here, we report the isolation of an AtPep1 receptor from the surface of Arabidopsis suspension-cultured cells. An (125)I-labeled AtPep1 analog interacted with suspension-cultured Arabidopsis with a K(d) of 0.25 nM, and an (125)I-labeled azido-Cys-AtPep1 photoaffinity analog specifically labeled a membrane-associated protein of approximately 170 kDa. The labeled protein was purified to homogeneity, and its tryptic peptides were identified as gene At1g73080, which encodes a leucine-rich repeat receptor kinase, here called PEPR1. Verification of the binding protein as the receptor for AtPep1 was established by demonstrating the loss of function of microsomal membranes of two SALK insertional mutants and by a gain in function of the alkalinization response to AtPep1 by tobacco suspension-cultured cells expressing the At1g73080 transgene. Synthetic homologs of AtPep1, deduced from the C termini of six known paralogs of PROPEP1, were biologically active and were competitors of the interaction of an AtPep1 radiolabeled analog with the receptor. The data are consistent with a role for PEPR1 as the receptor for AtPep1 to amplify innate immunity in response to pathogen attacks.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


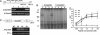
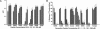
References
-
- Hoffman J. A., Kafatos F. C., Janeway C. A., Jr., Ezekowitz R. A. B. Science. 1999;284:1313–1318. - PubMed
-
- Menezes H., Jared C. Comp. Biochem. Physiol. C Toxicol. Pharmocol. 2002;132:1–7. - PubMed
-
- Nürnberger T., Brunner F., Kemmerling B., Piater L. Immunol. Rev. 2004;198:249–266. - PubMed
-
- Parker J. Trends Plant. Sci. 2003;8:245–247. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

