An endogenous peptide signal in Arabidopsis activates components of the innate immune response
- PMID: 16785434
- PMCID: PMC1502512
- DOI: 10.1073/pnas.0603727103
An endogenous peptide signal in Arabidopsis activates components of the innate immune response
Abstract
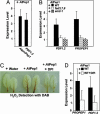
Innate immunity is initiated in animals and plants through the recognition of a variety of pathogen-associated molecules that in animals are called pathogen-associated molecular patterns and in plants are called elicitors. Some plant pathogen-derived elicitors have been identified as peptides, but peptide elicitors derived from the plant itself that activate defensive genes against pathogens have not been previously identified. Here, we report the isolation and characterization of a 23-aa peptide from Arabidopsis, called AtPep1, which activates transcription of the defensive gene defensin (PDF1.2) and activates the synthesis of H(2)O(2), both being components of the innate immune response. The peptide is derived from a 92-aa precursor encoded within a small gene that is inducible by wounding, methyl jasmonate, and ethylene. Constitutive expression of the AtPep1 precursor gene PROPEP1 in transgenic Arabidopsis plants causes a constitutive transcription of PDF1.2. When grown in soil, the transgenic plants exhibited an increased root development compared with WT plants and an enhanced resistance toward the root pathogen Pythium irregulare. Six paralogs of PROPEP1 are present in Arabidopsis, and orthologs have been identified in species of several agriculturally important plant families, where they are of interest for their possible use in crop improvement.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures




References
-
- Kamoun S. Curr. Opin. Plant Biol. 2001;4:295–300. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

