Comparative functional analysis of human medium-chain dehydrogenases, short-chain dehydrogenases/reductases and aldo-keto reductases with retinoids
- PMID: 16787387
- PMCID: PMC1570161
- DOI: 10.1042/BJ20051988
Comparative functional analysis of human medium-chain dehydrogenases, short-chain dehydrogenases/reductases and aldo-keto reductases with retinoids
Abstract
Retinoic acid biosynthesis in vertebrates occurs in two consecutive steps: the oxidation of retinol to retinaldehyde followed by the oxidation of retinaldehyde to retinoic acid. Enzymes of the MDR (medium-chain dehydrogenase/reductase), SDR (short-chain dehydrogenase/reductase) and AKR (aldo-keto reductase) superfamilies have been reported to catalyse the conversion between retinol and retinaldehyde. Estimation of the relative contribution of enzymes of each type was difficult since kinetics were performed with different methodologies, but SDRs would supposedly play a major role because of their low K(m) values, and because they were found to be active with retinol bound to CRBPI (cellular retinol binding protein type I). In the present study we employed detergent-free assays and HPLC-based methodology to characterize side-by-side the retinoid-converting activities of human MDR [ADH (alcohol dehydrogenase) 1B2 and ADH4), SDR (RoDH (retinol dehydrogenase)-4 and RDH11] and AKR (AKR1B1 and AKR1B10) enzymes. Our results demonstrate that none of the enzymes, including the SDR members, are active with CRBPI-bound retinoids, which questions the previously suggested role of CRBPI as a retinol supplier in the retinoic acid synthesis pathway. The members of all three superfamilies exhibit similar and low K(m) values for retinoids (0.12-1.1 microM), whilst they strongly differ in their kcat values, which range from 0.35 min(-1) for AKR1B1 to 302 min(-1) for ADH4. ADHs appear to be more effective retinol dehydrogenases than SDRs because of their higher kcat values, whereas RDH11 and AKR1B10 are efficient retinaldehyde reductases. Cell culture studies support a role for RoDH-4 as a retinol dehydrogenase and for AKR1B1 as a retinaldehyde reductase in vivo.
Figures


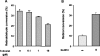
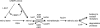
References
-
- Mangelsdorf D., Umesono K., Evans R. M. The retinoid receptors. In: Sporn M. B., Roberts A. B., Goodman D. S., editors. The Retinoids: Biology, Chemistry and Medicine. New York: Raven Press; 1994. pp. 319–350.
-
- Vogel S., Gamble M. V., Blaner W. S. Biosynthesis, absorption, metabolism and transport of retinoids. In: Nau H., Blaner W. S., editors. Retinoids: The Biochemical and Molecular Basis of Vitamin A and Retinoid Action, vol. 139. Berlin: Springer-Verlag; 1999. pp. 31–96.
-
- von Lintig J., Wyss A. Molecular analysis of vitamin A formation: cloning and characterization of β-carotene 15,15′-dioxygenases. Arch. Biochem. Biophys. 2001;385:47–52. - PubMed
-
- Napoli J. L. Interactions of retinoid binding proteins and enzymes in retinoid metabolism. Biochim. Biophys. Acta. 1999;1440:139–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

