Uracil salvage pathway in Lactobacillus plantarum: Transcription and genetic studies
- PMID: 16788187
- PMCID: PMC1483017
- DOI: 10.1128/JB.00195-06
Uracil salvage pathway in Lactobacillus plantarum: Transcription and genetic studies
Abstract
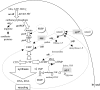
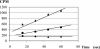
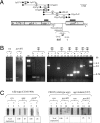
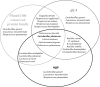
The uracil salvage pathway in Lactobacillus plantarum was demonstrated to be dependent on the upp-pyrP gene cluster. PyrP was the only high-affinity uracil transporter since a pyrP mutant no longer incorporated low concentrations of radioactively labeled uracil and had increased resistance to the toxic uracil analogue 5-fluorouracil. The upp gene encoded a uracil phosphoribosyltransferase (UPRT) enzyme catalyzing the conversion of uracil and 5-phosphoribosyl-alpha-1-pyrophosphate to UMP and pyrophosphate. Analysis of mutants revealed that UPRT is a major cell supplier of UMP synthesized from uracil provided by preformed nucleic acid degradation. In a mutant selection study, seven independent upp mutants were isolated and all were found to excrete low amounts of pyrimidines to the growth medium. Pyrimidine-dependent transcription regulation of the biosynthetic pyrimidine pyrR1-B-C-Aa1-Ab1-D-F-E operon was impaired in the upp mutants. Despite the fact that upp and pyrP are positioned next to each other on the chromosome, they are not cotranscribed. Whereas pyrP is expressed as a monocistronic message, the upp gene is part of the lp_2376-glyA-upp operon. The lp_2376 gene encodes a putative protein that belongs to the conserved protein family of translation modulators such as Sua5, YciO, and YrdC. The glyA gene encodes a putative hydroxymethyltransferase involved in C1 unit charging of tetrahydrofolate, which is required in the biosynthesis of thymidylate, pantothenate, and purines. Unlike upp transcription, pyrP transcription is regulated by exogenous pyrimidine availability, most likely by the same mechanism of transcription attenuation as that of the pyr operon.
Figures


References
-
- Andersen, P. S., J. M. Smith, and B. Mygind. 1992. Characterization of the upp gene encoding uracil phosphoribosyltransferase of Escherichia coli K12. Eur. J. Biochem. 204:51-56. - PubMed
-
- Beyer, N. H., P. Roepstorff, K. Hammer, and M. Kilstrup. 2003. Proteome analysis of the purine stimulon from Lactococcus lactis. Proteomics 3:786-797. - PubMed
-
- Bolivar, F., R. L. Rodriguez, M. C. Betlach, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene 2:75-93. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

