Integrated assessment and prediction of transcription factor binding
- PMID: 16789814
- PMCID: PMC1479087
- DOI: 10.1371/journal.pcbi.0020070
Integrated assessment and prediction of transcription factor binding
Abstract
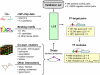
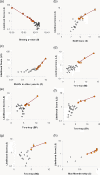
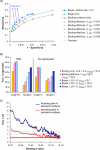
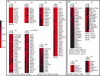
Systematic chromatin immunoprecipitation (chIP-chip) experiments have become a central technique for mapping transcriptional interactions in model organisms and humans. However, measurement of chromatin binding does not necessarily imply regulation, and binding may be difficult to detect if it is condition or cofactor dependent. To address these challenges, we present an approach for reliably assigning transcription factors (TFs) to target genes that integrates many lines of direct and indirect evidence into a single probabilistic model. Using this approach, we analyze publicly available chIP-chip binding profiles measured for yeast TFs in standard conditions, showing that our model interprets these data with significantly higher accuracy than previous methods. Pooling the high-confidence interactions reveals a large network containing 363 significant sets of factors (TF modules) that cooperate to regulate common target genes. In addition, the method predicts 980 novel binding interactions with high confidence that are likely to occur in so-far untested conditions. Indeed, using new chIP-chip experiments we show that predicted interactions for the factors Rpn4p and Pdr1p are observed only after treatment of cells with methyl-methanesulfonate, a DNA-damaging agent. We outline the first approach for consistently integrating all available evidences for TF-target interactions and we comprehensively identify the resulting TF module hierarchy. Prioritizing experimental conditions for each factor will be especially important as increasing numbers of chIP-chip assays are performed in complex organisms such as humans, for which "standard conditions" are ill defined.
Conflict of interest statement
Figures

References
-
- Pilpel Y, Sudarsanam P, Church GM. Identifying regulatory networks by combinatorial analysis of promotor elements. Nat Genet. 2001;29:153–159. - PubMed
-
- Bar-Joseph Z, Gerber GK, Lee TI, Rinaldi NJ, Yoo JY, et al. Computational discovery of gene modules and regulatory networks. Nat Biotechnol. 2003;21:1337–1342. - PubMed
-
- Reményi A, Scholer HR, Wilmanns M. Combinatorial control of gene expression. Nat Struct Mol Biol. 2004;11:812–815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

