A community resource benchmarking predictions of peptide binding to MHC-I molecules
- PMID: 16789818
- PMCID: PMC1475712
- DOI: 10.1371/journal.pcbi.0020065
A community resource benchmarking predictions of peptide binding to MHC-I molecules
Abstract
Recognition of peptides bound to major histocompatibility complex (MHC) class I molecules by T lymphocytes is an essential part of immune surveillance. Each MHC allele has a characteristic peptide binding preference, which can be captured in prediction algorithms, allowing for the rapid scan of entire pathogen proteomes for peptide likely to bind MHC. Here we make public a large set of 48,828 quantitative peptide-binding affinity measurements relating to 48 different mouse, human, macaque, and chimpanzee MHC class I alleles. We use this data to establish a set of benchmark predictions with one neural network method and two matrix-based prediction methods extensively utilized in our groups. In general, the neural network outperforms the matrix-based predictions mainly due to its ability to generalize even on a small amount of data. We also retrieved predictions from tools publicly available on the internet. While differences in the data used to generate these predictions hamper direct comparisons, we do conclude that tools based on combinatorial peptide libraries perform remarkably well. The transparent prediction evaluation on this dataset provides tool developers with a benchmark for comparison of newly developed prediction methods. In addition, to generate and evaluate our own prediction methods, we have established an easily extensible web-based prediction framework that allows automated side-by-side comparisons of prediction methods implemented by experts. This is an advance over the current practice of tool developers having to generate reference predictions themselves, which can lead to underestimating the performance of prediction methods they are not as familiar with as their own. The overall goal of this effort is to provide a transparent prediction evaluation allowing bioinformaticians to identify promising features of prediction methods and providing guidance to immunologists regarding the reliability of prediction tools.
Conflict of interest statement
Figures
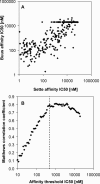

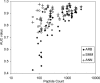
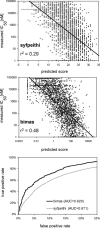
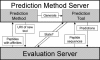
References
-
- Shastri N, Schwab S, Serwold T. Producing nature's gene-chips: The generation of peptides for display by MHC class I molecules. Annu Rev Immunol. 2002;20:463–493. - PubMed
-
- Marincola FM, Wang E, Herlyn M, Seliger B, Ferrone S. Tumors as elusive targets of T-cell–based active immunotherapy. Trends Immunol. 2003;24:335–342. - PubMed
-
- Descamps FJ, Van den Steen PE, Nelissen I, Van Damme J, Opdenakker G. Remnant epitopes generate autoimmunity: From rheumatoid arthritis and multiple sclerosis to diabetes. Adv Exp Med Biol. 2003;535:69–77. - PubMed
-
- Bhasin M, Raghava GP. SVM based method for predicting HLA-DRB1*0401 binding peptides in an antigen sequence. Bioinformatics. 2004;20:421–423. - PubMed
-
- Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, et al. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304–314. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

