The life-cycle of operons
- PMID: 16789824
- PMCID: PMC1480536
- DOI: 10.1371/journal.pgen.0020096
The life-cycle of operons
Erratum in
- PLoS Genet. 2006 Jul;2(7):e126
Abstract
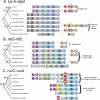
Operons are a major feature of all prokaryotic genomes, but how and why operon structures vary is not well understood. To elucidate the life-cycle of operons, we compared gene order between Escherichia coli K12 and its relatives and identified the recently formed and destroyed operons in E. coli. This allowed us to determine how operons form, how they become closely spaced, and how they die. Our findings suggest that operon evolution may be driven by selection on gene expression patterns. First, both operon creation and operon destruction lead to large changes in gene expression patterns. For example, the removal of lysA and ruvA from ancestral operons that contained essential genes allowed their expression to respond to lysine levels and DNA damage, respectively. Second, some operons have undergone accelerated evolution, with multiple new genes being added during a brief period. Third, although genes within operons are usually closely spaced because of a neutral bias toward deletion and because of selection against large overlaps, genes in highly expressed operons tend to be widely spaced because of regulatory fine-tuning by intervening sequences. Although operon evolution may be adaptive, it need not be optimal: new operons often comprise functionally unrelated genes that were already in proximity before the operon formed.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures



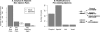
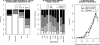

References
-
- Wolf Y, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–72. - PubMed
-
- de Daruvar A, Collado-Vides J, Valencia A. Analysis of the cellular functions of Escherichia coli operons and their conservation in Bacillus subtilis. J Mol Evol. 2002;55:211–221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

